4MAF
 
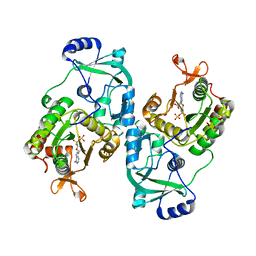 | | Soybean ATP Sulfurylase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase | | Authors: | Herrmann, J, Ravilious, G.E, McKinney, S.E, Westfall, C.S, Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation.
J.Biol.Chem., 289, 2014
|
|
5CB8
 
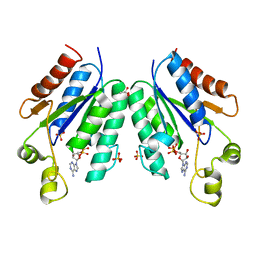 | |
5CB6
 
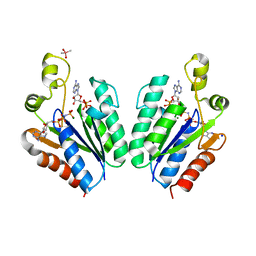 | | Structure of adenosine-5'-phosphosulfate kinase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Herrmann, J, Jez, J.M. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Recapitulating the Structural Evolution of Redox Regulation in Adenosine 5'-Phosphosulfate Kinase from Cyanobacteria to Plants.
J.Biol.Chem., 290, 2015
|
|
6P5T
 
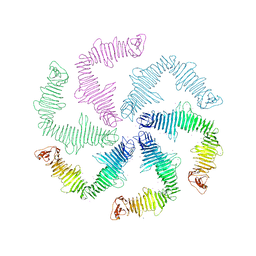 | | Surface-layer (S-layer) RsaA protein from Caulobacter crescentus bound to strontium and iodide | | Descriptor: | IODIDE ION, S-layer protein, STRONTIUM ION | | Authors: | Chan, A.C, Herrmann, J, Smit, J, Wakatsuki, S, Murphy, M.E. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A bacterial surface layer protein exploits multistep crystallization for rapid self-assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7O2L
 
 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
4EQL
 
 | | Crystal Structure of GH3.12 in complex with AMP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | Authors: | Westfall, C, Zubieta, C, Nanao, M, Herrmann, J, Jez, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EPM
 
 | | Crystal Structure of Arabidopsis GH3.12 (PBS3) in Complex with AMP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EPL
 
 | | Crystal Structure of Arabidopsis thaliana GH3.11 (JAR1) in Complex with JA-Ile | | Descriptor: | Jasmonic acid-amido synthetase JAR1, N-({(1R,2R)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
1G3F
 
 | | NMR STRUCTURE OF A 9 RESIDUE PEPTIDE FROM SMAC/DIABLO COMPLEXED TO THE BIR3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN 3, SMAC, ZINC ION | | Authors: | Liu, Z, Sun, C, Olejniczak, E.T, Meadows, R.P, Betz, S.F, Oost, T, Herrmann, J, Wu, J.C, Fesik, S.W. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain.
Nature, 408, 2000
|
|
8B7Y
 
 | | Cryo-EM structure of the E.coli 70S ribosome in complex with the antibiotic Myxovalargin B. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Koller, T.O, Graf, M, Wilson, D.N. | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-25 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
8OET
 
 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
5O28
 
 | | E. coli FolD apo | | Descriptor: | Bifunctional protein FolD | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-05-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD.
Nat Commun, 8, 2017
|
|
5O2A
 
 | | FolD Q98H | | Descriptor: | Bifunctional protein FolD | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-05-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD.
Nat Commun, 8, 2017
|
|
5O22
 
 | | E. coli FolD in complex with carolacton | | Descriptor: | Bifunctional protein FolD, Carolacton | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-05-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD.
Nat Commun, 8, 2017
|
|
6P3Q
 
 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
8ADG
 
 | | Cryo-EM structure of Darobactin 22 bound BAM complex | | Descriptor: | Darobactin 22, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ADI
 
 | | Cryo-EM structure of Darobactin 9 bound BAM complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6VVY
 
 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with Sorangicin | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VW0
 
 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVX
 
 | | Mycobacterium tuberculosis WT RNAP transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVZ
 
 | | Mycobacterium tuberculosis RNAP S456L mutant transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (62-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4EWV
 
 | | Crystal structure of GH3.12 in complex with AMPCPP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Zubieta, C, Nanao, M, Jez, J. | | Deposit date: | 2012-04-28 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
6VVT
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVV
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVS
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Braffman, N, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
