1MRN
 
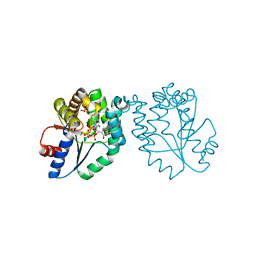 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH BISUBSTRATE INHIBITOR (TP5A) | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
1MRS
 
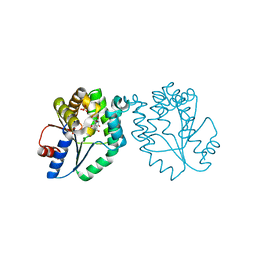 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH 5-CH2OH DEOXYURIDINE MONOPHOSPHATE | | Descriptor: | 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
8Q5P
 
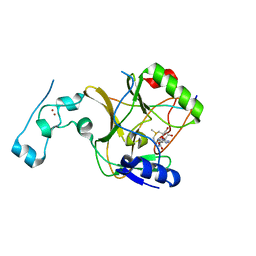 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
6Q42
 
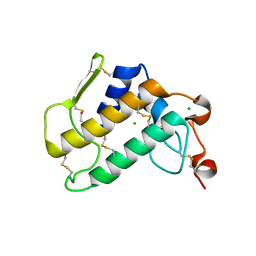 | | Crystal Structure of Human Pancreatic Phospholipase A2 | | Descriptor: | CHLORIDE ION, Phospholipase A2 | | Authors: | Saul, F, Haouz, A, Lambeau, G, Theze, J. | | Deposit date: | 2018-12-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PLA2G1B is involved in CD4 anergy and CD4 lymphopenia in HIV-infected patients.
J.Clin.Invest., 130, 2020
|
|
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4USS
 
 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
5IKP
 
 | | Crystal structure of human brain glycogen phosphorylase bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, brain form, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
5IKO
 
 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
2UYL
 
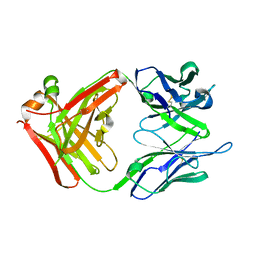 | | Crystal structure of a monoclonal antibody directed against an antigenic determinant common to Ogawa and Inaba serotypes of Vibrio cholerae O1 | | Descriptor: | MONOCLONAL ANTIBODY F-22-30 | | Authors: | Ahmed, F, Haouz, A, Nato, F, Fournier, J.M, Alzari, P.M. | | Deposit date: | 2007-04-10 | | Release date: | 2008-05-13 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monoclonal Antibody Directed Against an Antigenic Determinant Common to Ogawa and Inaba Serotypes of Vibrio Cholerae O1.
Proteins, 70, 2008
|
|
2CKD
 
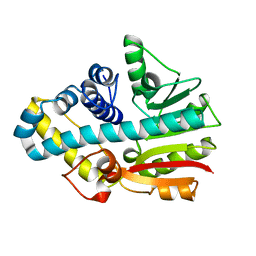 | | Crystal structure of ML2640 from Mycobacterium leprae | | Descriptor: | PUTATIVE S-ADENOSYL-L-METHIONINE-DEPENDENT METHYLTRANSFERASE ML2640 | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Alzari, P.M. | | Deposit date: | 2006-04-17 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
6HJF
 
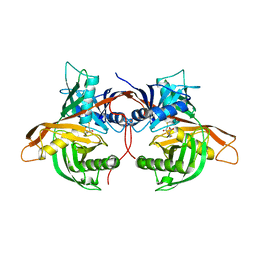 | | Trypanosoma cruzi proline racemase in complex with inhibitor BrOxoPA | | Descriptor: | LAEVULINIC ACID, Proline racemase A | | Authors: | Saul, F, Haouz, A, Uriac, P, Blondel, A, Minoprio, P. | | Deposit date: | 2018-09-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed mono- and di-covalent inhibitors trap modeled functional motions for Trypanosoma cruzi proline racemase in crystallography.
PLoS Negl Trop Dis, 12, 2018
|
|
6HJE
 
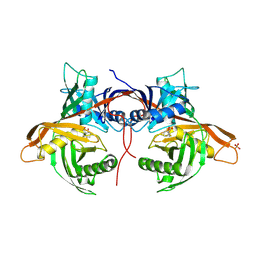 | | Trypanosoma cruzi proline racemase in complex with inhibitor NG-P27 | | Descriptor: | 2-[(1~{S})-2-oxidanylidenecyclopentyl]ethanoic acid, PHOSPHATE ION, Proline racemase A | | Authors: | Saul, F, Haouz, A, Uriac, P, Blondel, A, Minoprio, P. | | Deposit date: | 2018-09-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed mono- and di-covalent inhibitors trap modeled functional motions for Trypanosoma cruzi proline racemase in crystallography.
PLoS Negl Trop Dis, 12, 2018
|
|
7BL7
 
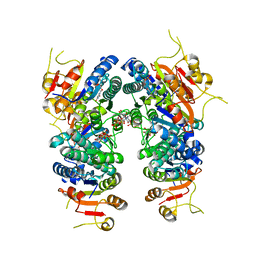 | | Crystal structure of UMPK from M. tuberculosis in complex with UDP and UTP (P21212 form) | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Walter, P, Labesse, G, Haouz, A, Mechaly, A.E, Munier-Lehmann, H. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
7BIX
 
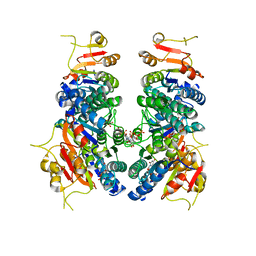 | | Crystal structure of UMPK from M. tuberculosis in complex with UDP and UTP (C2 form) | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Labesse, G, Walter, P, Haouz, A, Mechaly, A.E, Munier-Lehmann, H. | | Deposit date: | 2021-01-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
8QKE
 
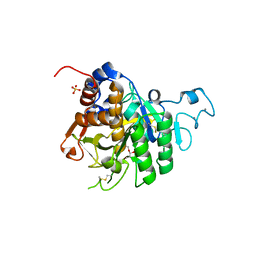 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MH-13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MH-13), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKG
 
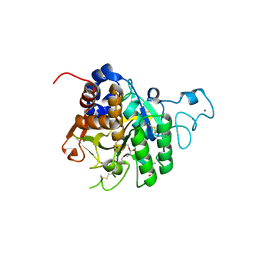 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-125) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-125), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
8QKJ
 
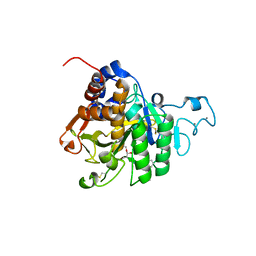 | | PvSub1 Catalytic Domain in Complex with Peptidomimetic Inhibitor (MAM-133) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Peptidomimetic Inhibitor (MAM-133), ... | | Authors: | Batista, F.A, Martinez, M, Bouillon, A, Mechaly, A, Alzari, P.M, Haouz, A, Barale, J.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Insights from structure-activity relationships and the binding mode of peptidic alpha-ketoamide inhibitors of the malaria drug target subtilisin-like SUB1.
Eur.J.Med.Chem., 269, 2024
|
|
4YIN
 
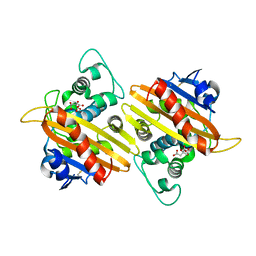 | | Crystal structure of the extended-spectrum beta-lactamase OXA-145 | | Descriptor: | Beta-lactamase, CITRATE ANION | | Authors: | Meziane-Cherif, D, Bonnet, R, Haouz, A, Courvalin, P. | | Deposit date: | 2015-03-02 | | Release date: | 2016-02-10 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the loss of penicillinase and the gain of ceftazidimase activities by OXA-145 beta-lactamase in Pseudomonas aeruginosa.
J. Antimicrob. Chemother., 71, 2016
|
|
5AHN
 
 | | IMP-bound form of the D199N mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, MAGNESIUM ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AHL
 
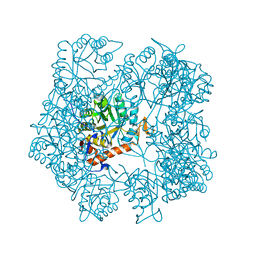 | | Apo-form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, SODIUM ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2CDN
 
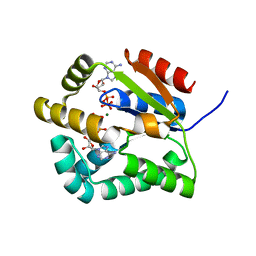 | | Crystal structure of Mycobacterium tuberculosis adenylate kinase complexed with two molecules of ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE, MAGNESIUM ION | | Authors: | Bellinzoni, M, Haouz, A, Grana, M, Munier-Lehmann, H, Alzari, P.M. | | Deposit date: | 2006-01-25 | | Release date: | 2006-05-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Adenylate Kinase in Complex with Two Molecules of Adp and Mg2+ Supports an Associative Mechanism for Phosphoryl Transfer.
Protein Sci., 15, 2006
|
|
2FWV
 
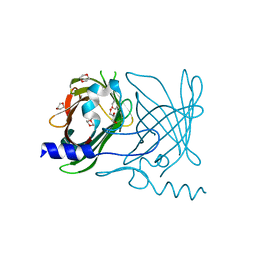 | | Crystal Structure of Rv0813 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, hypothetical protein MtubF_01000852 | | Authors: | Shepard, W, Haouz, A, Grana, M, Buschiazzo, A, Betton, J.M, Cole, S.T, Alzari, P.M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-03 | | Release date: | 2006-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Rv0813c from Mycobacterium tuberculosis Reveals a New Family of Fatty Acid-Binding Protein-Like Proteins in Bacteria
J.Bacteriol., 189, 2007
|
|
6MFV
 
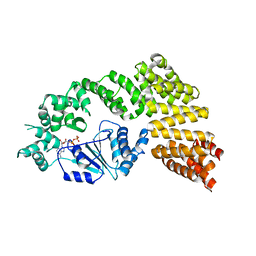 | | Crystal structure of the Signal Transduction ATPase with Numerous Domains (STAND) protein with a tetratricopeptide repeat sensor PH0952 from Pyrococcus horikoshii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, tetratricopeptide repeat sensor PH0952 | | Authors: | Lisa, M.N, Alzari, P.M, Haouz, A, Danot, O. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor.
Nucleic Acids Res., 47, 2019
|
|
8OEP
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HPV18 E6 oncoprotein C-terminal peptide | | Descriptor: | Protein E6, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | Deposit date: | 2023-03-11 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
5AHM
 
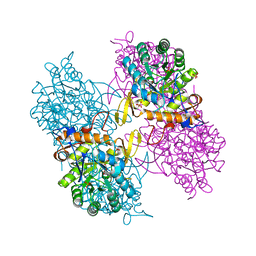 | | IMP-bound form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, PHOSPHATE ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
