8J5I
 
 | | MALT1 death domain | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Lo, Y.C, Kuo, B.J. | | Deposit date: | 2023-04-22 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MALT1 death domain
To Be Published
|
|
5SUM
 
 | | Ribosome assembly factor NSA1 | | Descriptor: | Ribosome biogenesis protein NSA1 | | Authors: | Lo, Y.H, Stanley, R.E. | | Deposit date: | 2016-08-03 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ribosome assembly factor
To Be Published
|
|
5SUI
 
 | |
1V2G
 
 | | The L109P mutant of E. coli Thioesterase I/Protease I/Lysophospholipase L1 (TAP) in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, IMIDAZOLE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2003-10-15 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1J00
 
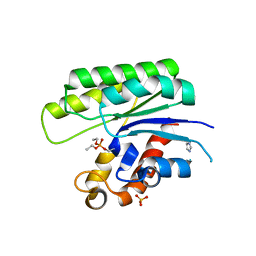 | |
1IVN
 
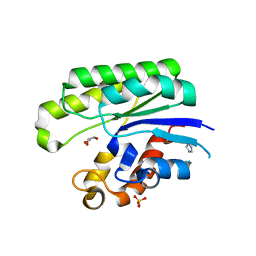 | | E.coli Thioesterase I/Protease I/Lysophospholiase L1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioesterase I | | Authors: | Lo, Y.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
4APV
 
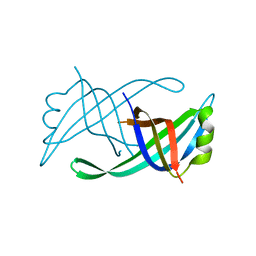 | | The Klebsiella pneumoniae primosomal PriB protein: identification, crystal structure, and ssDNA binding mode | | Descriptor: | PRIMOSOMAL REPLICATION PROTEIN N | | Authors: | Lo, Y.H, Huang, Y.H, Hsiao, C.D, Huang, C.Y. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal Structure and DNA-Binding Mode of Klebsiella Pneumoniae Primosomal Prib Protein.
Genes Cells, 17, 2012
|
|
4HIV
 
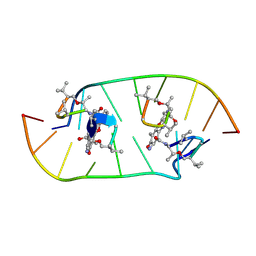 | | Structure of actinomycin D d(ATGCGGCAT) complex | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*AP*T)-3') | | Authors: | Lo, Y.S, Tseng, W.H, Hou, M.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats.
Nucleic Acids Res., 41, 2013
|
|
5H07
 
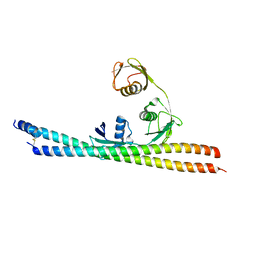 | | TNIP2-Ub complex, C2 form | | Descriptor: | Polyubiquitin-C, TNFAIP3-interacting protein 2 | | Authors: | Lo, Y.C, Lin, S.C. | | Deposit date: | 2016-10-04 | | Release date: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structural Insights into Linear Tri-ubiquitin Recognition by A20-Binding Inhibitor of NF-kappa B, ABIN-2
Structure, 25, 2017
|
|
8RJK
 
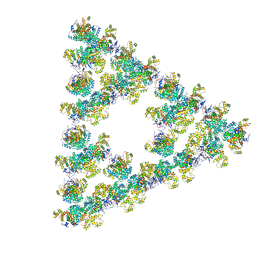 | | Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 2024
|
|
2VYF
 
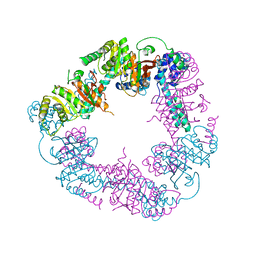 | | Crystal Structure of the DnaC | | Descriptor: | GOLD ION, REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
2VYE
 
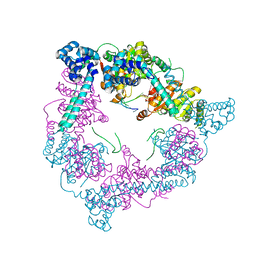 | | Crystal Structure of the DnaC-ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP)-3', REPLICATIVE DNA HELICASE | | Authors: | Lo, Y.H, Tsai, K.L, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | The Crystal Structure of a Replicative Hexameric Helicase Dnac and its Complex with Single-Stranded DNA.
Nucleic Acids Res., 37, 2009
|
|
1U8U
 
 | | E. coli Thioesterase I/Protease I/Lysophospholiase L1 in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, GLYCEROL, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2004-08-07 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
7EAO
 
 | |
7EAL
 
 | |
7EB9
 
 | |
1JRL
 
 | | Crystal structure of E. coli Lysophospholiase L1/Acyl-CoA Thioesterase I/Protease I L109P mutant | | Descriptor: | Acyl-CoA Thioesterase I, IMIDAZOLE, SULFATE ION | | Authors: | Lo, Y.-C, Lin, S.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2001-08-14 | | Release date: | 2003-07-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
3FX0
 
 | |
8AN1
 
 | |
8BEI
 
 | |
2ZZB
 
 | | Crystal structure of human thioredoxin reductase I and terpyridine platinum(II) | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Lo, Y.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2009-02-09 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Terpyridine-platinum(II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase 1.
J.Inorg.Biochem., 103, 2009
|
|
2ZZ0
 
 | | Crystal structure of human thioredoxin reductase I (SeCys 498 Cys) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thioredoxin reductase 1, ... | | Authors: | Lo, Y.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Terpyridine-platinum(II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase 1.
J.Inorg.Biochem., 103, 2009
|
|
2ZZC
 
 | | Crystal structure of NADP(H):human thioredoxin reductase I | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Lo, Y.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2009-02-09 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Terpyridine-platinum(II) complexes are effective inhibitors of mammalian topoisomerases and human thioredoxin reductase 1.
J.Inorg.Biochem., 103, 2009
|
|
3RNZ
 
 | |
3RO1
 
 | |
