6QG1
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 2) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG6
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model D) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG2
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model A) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG0
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 1) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG3
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model B) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG5
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5IQR
 
 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
3D36
 
 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
5MDW
 
 | | Structure of ArfA(A18T) and RF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDV
 
 | | Structure of ArfA and RF2 bound to the 70S ribosome (accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDY
 
 | | Structure of ArfA and TtRF2 bound to the 70S ribosome (pre-accommodated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5MDZ
 
 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
6YBT
 
 | | Structure of a human 48S translational initiation complex - eIF3bgi | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBV
 
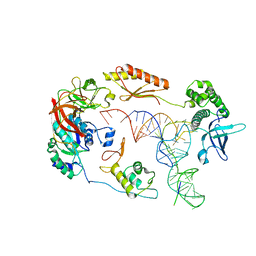 | | Structure of a human 48S translational initiation complex - eIF2-TC | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBS
 
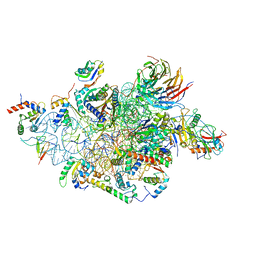 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBW
 
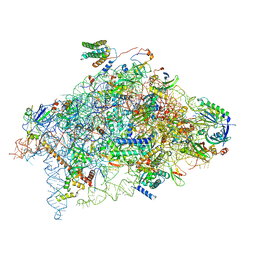 | | Structure of a human 48S translational initiation complex - 40S body | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBD
 
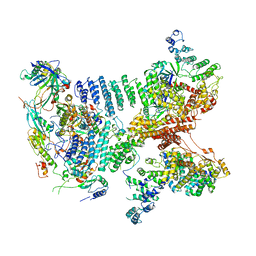 | | Structure of a human 48S translational initiation complex - eIF3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14, 40S ribosomal protein S17, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6FYX
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6GSM
 
 | | Structure of a partial yeast 48S preinitiation complex in open conformation. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-06-14 | | Release date: | 2019-07-31 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Large-scale movement of eIF3 domains during translation initiation modulate start codon selection.
Nucleic Acids Res., 2021
|
|
4BKX
 
 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
4CYJ
 
 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
4CYI
 
 | | Chaetomium thermophilum Pan3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
