6CSE
 
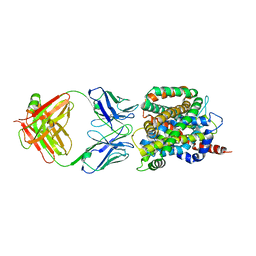 | | Crystal structure of sodium/alanine symporter AgcS with L-alanine bound | | Descriptor: | ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
9EAM
 
 | |
2LI8
 
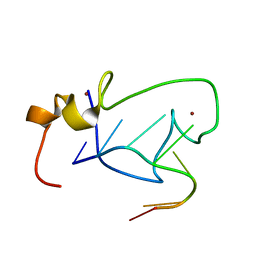 | |
3BWA
 
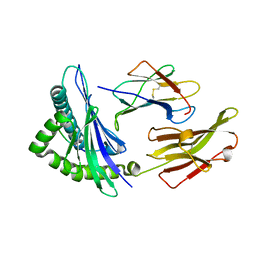 | | Crystal Structure of HLA B*3508 in complex with a HCMV 8-mer peptide from the pp65 protein | | Descriptor: | Beta-2-microglobulin, FPT peptide from 65 kDa lower matrix phosphoprotein, HLA class I histocompatibility antigen, ... | | Authors: | Wynn, K.K, Marland, Z, Cooper, L, Silins, S.L, Gras, S, Archbold, J.K, Tynan, F.E, Miles, J.J, McCluskey, J, Burrows, S.R, Rossjohn, J, Khanna, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Blood, 111, 2008
|
|
5AMT
 
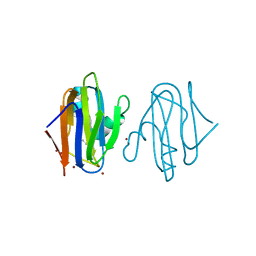 | | Intracellular growth locus protein E | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Intracellular Growth Locus E (Igle) Protein from Francisella Tularensis Subsp. Novicida.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2AK4
 
 | | Crystal Structure of SB27 TCR in complex with HLA-B*3508-13mer peptide | | Descriptor: | Beta-2-microglobulin, EBV peptide LPEPLPQGQLTAY, HLA-B35 variant, ... | | Authors: | Tynan, F.E, Burrows, S.R, Buckle, A.M, Clements, C.S, Borg, N.A, Miles, J.J, Beddoe, T, Whisstock, J.C, Wilce, M.C, Silins, S.L, Burrows, J.M, Kjer-Nielsen, L, Konstenko, L, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor recognition of a 'super-bulged' major histocompatibility complex class I-bound peptide
Nat.Immunol., 6, 2005
|
|
2AXF
 
 | | The Immunogenicity of a Viral Cytotoxic T Cell Epitope is controlled by its MHC-bound Conformation | | Descriptor: | 10-mer peptide from BZLF1 trans-activator protein, ACETIC ACID, Beta-2-microglobulin, ... | | Authors: | Tynan, F.E, Elhassen, D, Purcell, A.W, Burrows, J.M, Borg, N.A, Miles, J.J, Williamson, N.A, Green, K.J, Tellam, J, Kjer-Nielsen, L, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation
J.Exp.Med., 202, 2005
|
|
1IU5
 
 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IU6
 
 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2SEM
 
 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PROTEIN (SEX MUSCLE ABNORMAL PROTEIN 5), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
5J1S
 
 | | TorsinA-LULL1 complex, H. sapiens, bound to VHH-BS2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Demircioglu, F.E, Schwartz, T.U. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia.
Elife, 5, 2016
|
|
5HCL
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with DMA | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N},~{N}-dimethylethanamide | | Authors: | Dong, J, Weber, F.E, Caflisch, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N,N Dimethylacetamide a drug excipient that acts as bromodomain ligand for osteoporosis treatment.
Sci Rep, 7, 2017
|
|
8TAH
 
 | | Cryo-EM structure of Cortactin-bound to Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1A, ... | | Authors: | Fregoso, F.E, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of synergistic activation of Arp2/3 complex by cortactin and WASP-family proteins.
Nat Commun, 14, 2023
|
|
5I9S
 
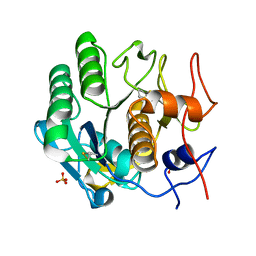 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
8T7R
 
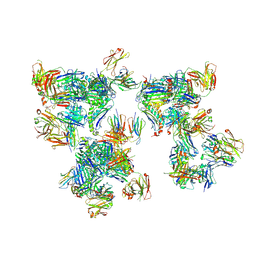 | | Crystal structure of human leukocyte antigen A*0101 in complex with the Fab of alloreactive antibody E07 | | Descriptor: | Beta-2-microglobulin, Fab heavy chain from antibody JTK191b E07, Light chain from antibody JTK191b E07, ... | | Authors: | Green, T.J, Killian Jr, J.T, Qiu, S, Macon, K.J, Yang, G, King, R.G, Lund, F.E. | | Deposit date: | 2023-06-21 | | Release date: | 2024-12-25 | | Last modified: | 2026-01-14 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Topography of the HLA-A protein enforces shared and convergent immunodominant B cell and antibody alloresponses in transplant recipients.
Immunity, 58, 2025
|
|
8T6M
 
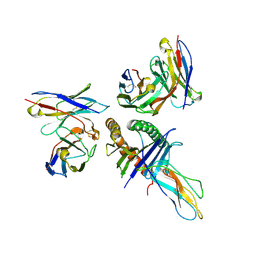 | | Human leukocyte antigen bound by two alloreactive antibody Fabs | | Descriptor: | Beta-2-microglobulin, HLA_A0101_peptide, JTK191b_L02_Fab, ... | | Authors: | Kizziah, J.L, Killian, J.T, Diaz-Avalos, R, Qiu, S, Yang, G, Green, T, Lund, F.E. | | Deposit date: | 2023-06-16 | | Release date: | 2024-12-25 | | Last modified: | 2026-01-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Topography of the HLA-A protein enforces shared and convergent immunodominant B cell and antibody alloresponses in transplant recipients.
Immunity, 58, 2025
|
|
9B9X
 
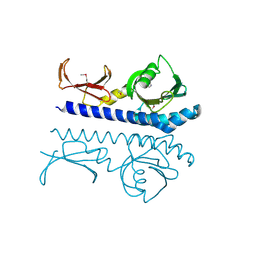 | | Crystal structure of the ligand binding domain of the Halomonas titanicae chemoreceptor Htc10 in complex with hypoxanthine | | Descriptor: | Chemotaxis protein, HYPOXANTHINE | | Authors: | Ramos Ricciuti, F.E, Herrera Seitz, M.K, Gasperotti, A.F, Boyko, A, Jung, K, Bellinzoni, M, Lisa, M.N, Studdert, C.A. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | The chemoreceptor controlling the Wsp-like transduction pathway in Halomonas titanicae KHS3 binds and responds to purine derivatives.
Febs J., 292, 2025
|
|
9BA3
 
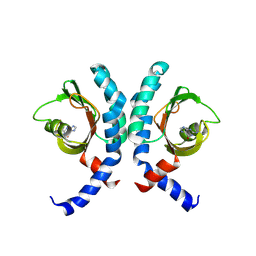 | | High-resolution crystal structure of the ligand binding domain of the Halomonas titanicae chemoreceptor Htc10 in complex with guanine | | Descriptor: | Chemotaxis protein, GUANINE | | Authors: | Ramos Ricciuti, F.E, Herrera Seitz, M.K, Gasperotti, A.F, Boyko, A, Jung, K, Bellinzoni, M, Lisa, M.N, Studdert, C.A. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The chemoreceptor controlling the Wsp-like transduction pathway in Halomonas titanicae KHS3 binds and responds to purine derivatives.
Febs J., 292, 2025
|
|
9B9S
 
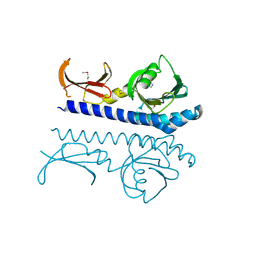 | | Crystal structure of the ligand binding domain of the Halomonas titanicae chemoreceptor Htc10 in complex with guanine | | Descriptor: | Chemotaxis protein, GUANINE | | Authors: | Ramos Ricciuti, F.E, Herrera Seitz, M.K, Gasperotti, A.F, Boyko, A, Jung, K, Bellinzoni, M, Lisa, M.N, Studdert, C.A. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The chemoreceptor controlling the Wsp-like transduction pathway in Halomonas titanicae KHS3 binds and responds to purine derivatives.
Febs J., 292, 2025
|
|
6OIF
 
 | | Cryo-EM structure of human TorsinA filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Torsin-1A | | Authors: | Zheng, W, Demircioglu, F.E, Schwartz, T.U, Egelman, E.H. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The AAA + ATPase TorsinA polymerizes into hollow helical tubes with 8.5 subunits per turn.
Nat Commun, 10, 2019
|
|
2R0N
 
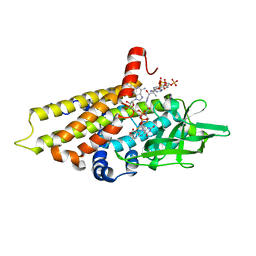 | | The effect of a Glu370Asp mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 3-thiaglutaryl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Albro, M, Fu, Z, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
4ER4
 
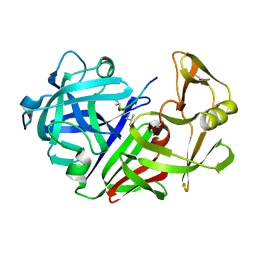 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
4FRN
 
 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | Descriptor: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
