5FFO
 
 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5FFG
 
 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
3GB8
 
 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
1Z9M
 
 | | Crystal Structure of Nectin-like molecule-1 protein Domain 1 | | Descriptor: | GAPA225 | | Authors: | Dong, X, Xu, F, Gong, Y, Gao, J, Lin, P, Chen, T, Peng, Y, Qiang, B, Yuan, J, Peng, X, Rao, Z. | | Deposit date: | 2005-04-03 | | Release date: | 2006-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the V Domain of Human Nectin-like Molecule-1/Syncam3/Tsll1/Igsf4b, a Neural Tissue-specific Immunoglobulin-like Cell-Cell Adhesion Molecule
J.Biol.Chem., 281, 2006
|
|
1WQL
 
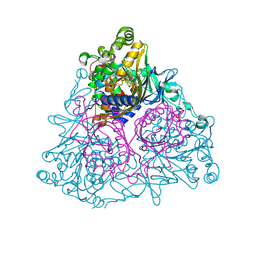 | | Cumene dioxygenase (cumA1A2) from Pseudomonas fluorescens IP01 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, OXYGEN MOLECULE, ... | | Authors: | Dong, X, Fushinobu, S, Fukuda, E, Terada, T, Nakamura, S, Shimizu, K, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Cumene Dioxygenase from Pseudomonas fluorescens IP01
J.BACTERIOL., 187, 2005
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | Descriptor: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | Authors: | Dong, X, Gong, Z, Hu, Y.F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
2DUD
 
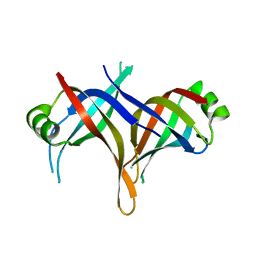 | |
2EGV
 
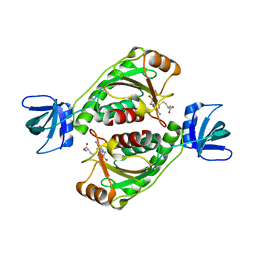 | |
2EGW
 
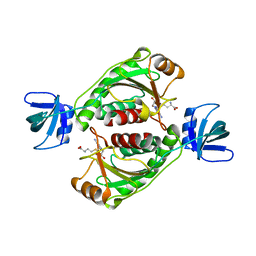 | |
1VS3
 
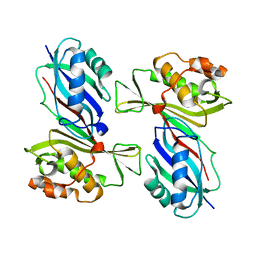 | | Crystal Structure of the tRNA Pseudouridine Synthase TruA From Thermus thermophilus HB8 | | Descriptor: | tRNA pseudouridine synthase A | | Authors: | Dong, X, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tRNA pseudouridine synthase TruA from Thermus thermophilus HB8.
Rna Biol., 3, 2006
|
|
2MVU
 
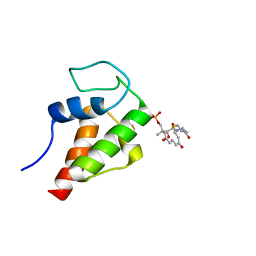 | | Solution Structure of the 3,7-dioxo-octyl Actinorhodin Acyl Carrier Protein from Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N~3~-{(2S)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-{2-[(3,7-dioxooctyl)sulfanyl]ethyl}-beta-alaninamide | | Authors: | Dong, X, Bailey, C, Williams, C, Crosby, J, Simpson, T.J, Willis, C.L, Crump, M.P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Method: | SOLUTION NMR | | Cite: | ACP-ligand recognition: Selection of derivatized aromatic biosynthetic intermediates
To be Published
|
|
2MVV
 
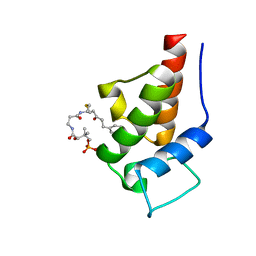 | | Solution Structure of the 5-phenyl-3-oxo-pentyl Actinorhodin Acyl Carrier Protein from Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N~3~-{(2S)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-{2-[(3-oxo-5-phenylpentyl)sulfanyl]ethyl}-beta-alaninamide | | Authors: | Dong, X, Bailey, C, Williams, C, Crosby, J, Simpson, T.J, Willis, C.L, Crump, M.P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Method: | SOLUTION NMR | | Cite: | ACP-ligand recognition: Selection of derivatized aromatic biosynthetic intermediates
To be Published
|
|
4G1E
 
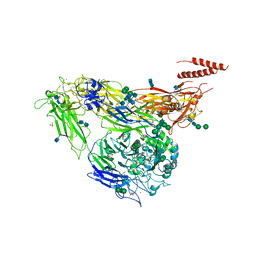 | | Crystal structure of integrin alpha V beta 3 with coil-coiled tag. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Mi, L, Zhu, J, Wang, W, Luo, B, Springer, T.A. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaV Beta3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4UM9
 
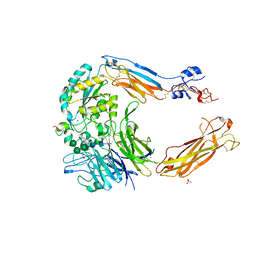 | | Crystal structure of alpha V beta 6 with peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UM8
 
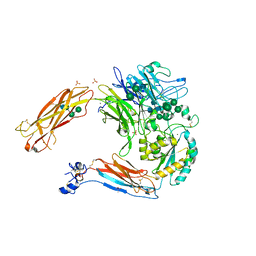 | | Crystal structure of alpha V beta 6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Nat.Struct.Mol.Biol., 21, 2014
|
|
7CYZ
 
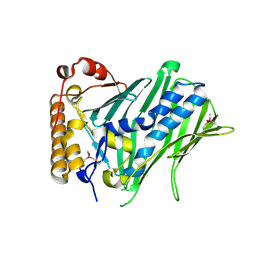 | | The structure of human ORP3 OSBP-related domain | | Descriptor: | Oxysterol-binding protein-related protein 3 | | Authors: | Dong, X. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of ORP3 reveals the conservative PI4P binding pattern.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
5XK4
 
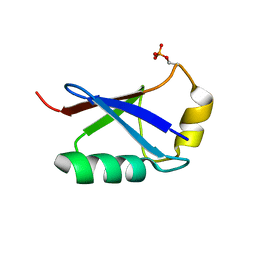 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Z2T
 
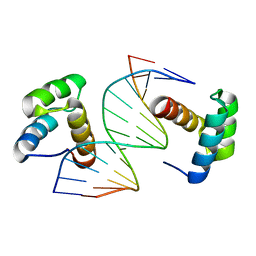 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
6A8R
 
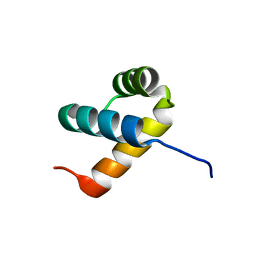 | | Crystal structure of DUX4 HD2 domain associated with ERG DNA binding site | | Descriptor: | DNA (5'-D(P*AP*AP*TP*CP*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*TP*GP*AP*GP*AP*TP*T)-3'), Double homeobox protein 4 | | Authors: | Dong, X, Zhang, H, Cheng, N, Meng, G. | | Deposit date: | 2018-07-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DUX4HD2-DNAERGstructure reveals new insight into DUX4-Responsive-Element.
Leukemia, 33, 2019
|
|
6N29
 
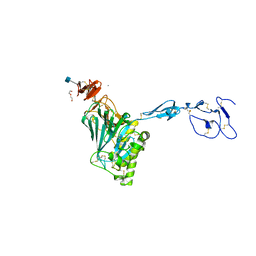 | | Crystal structure of monomeric von Willebrand Factor D`D3 assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, X, Arndt, J.W, Springer, T.A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis.
Blood, 133, 2019
|
|
2L22
 
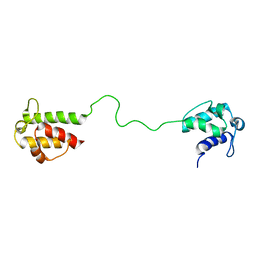 | | Mupirocin didomain ACP | | Descriptor: | Mupirocin didomain Acyl Carrier Protein | | Authors: | Dong, X, Williams, C, Crump, M.P, Wattana-amorn, P. | | Deposit date: | 2010-08-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif flags acyl carrier proteins for beta-branching in polyketide synthesis.
Nat.Chem.Biol., 9, 2013
|
|
3EBM
 
 | |
7LMX
 
 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
