1H8X
 
 | | Domain-swapped Dimer of a Human Pancreatic Ribonuclease Variant | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Canals, A, Pous, J, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2001-02-16 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of an Engineered Domain-Swapped Ribonuclease Dimer and its Implications for the Evolution of Proteins Toward Oligomerization
Structure, 9, 2001
|
|
3L0O
 
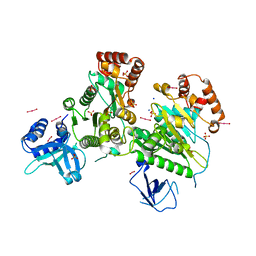 | | Structure of RNA-free Rho transcription termination factor from Thermotoga maritima | | Descriptor: | SODIUM ION, SULFATE ION, Transcription termination factor rho, ... | | Authors: | Canals, A, Uson, I, Coll, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture
J.Mol.Biol., 400, 2010
|
|
1NQ6
 
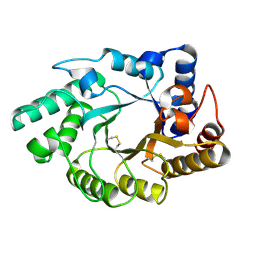 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4MNB
 
 | | Crystal Structure of a complex between the marine anticancer drug Variolin B and DNA | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, COBALT (II) ION, ... | | Authors: | Canals, A, Arribas-Bosacoma, R, Alvarez, M, Albericio, F, Aymami, J, Coll, M. | | Deposit date: | 2013-09-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Intercalative DNA binding of the marine anticancer drug variolin B.
Sci Rep, 7, 2017
|
|
1Z3F
 
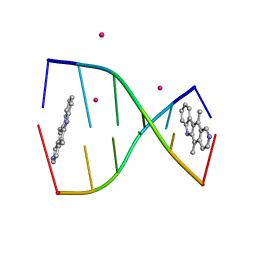 | | Structure of ellipticine in complex with a 6-bp DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', COBALT (II) ION, ELLIPTICINE | | Authors: | Canals, A, Purciolas, M, Aymami, J, Coll, M. | | Deposit date: | 2005-03-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8B4C
 
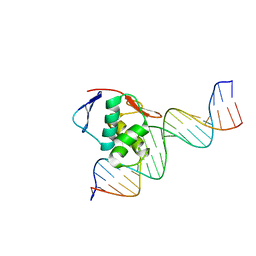 | | ToxR bacterial transcriptional regulator bound to 20 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (20-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4B
 
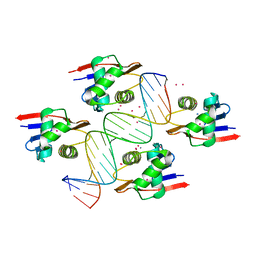 | | ToxR bacterial transcriptional regulator bound to 19 bp ompU promoter DNA | | Descriptor: | AMMONIUM ION, CADMIUM ION, Cholera toxin transcriptional activator, ... | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4E
 
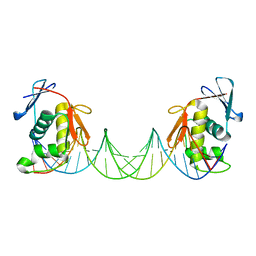 | | ToxR bacterial transcriptional regulator bound to 25 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (25-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4D
 
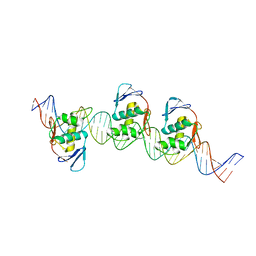 | | ToxR bacterial transcriptional regulator bound to 40 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (40-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2ERM
 
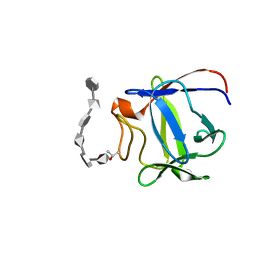 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
3O76
 
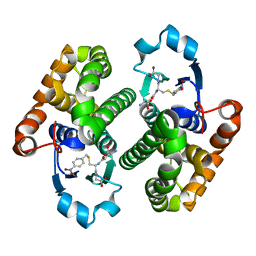 | |
3T72
 
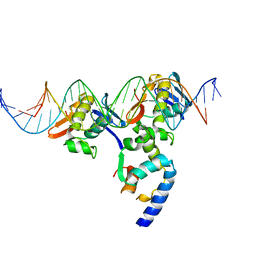 | | PhoB(E)-Sigma70(4)-(RNAP-Betha-flap-tip-helix)-DNA Transcription Activation Sub-Complex | | Descriptor: | PHO BOX DNA (STRAND 1), PHO BOX DNA (STRAND 2), Phosphate regulon transcriptional regulatory protein phoB, ... | | Authors: | Blanco, A.G, Canals, A, Bernues, J, Sola, M, Coll, M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.33 Å) | | Cite: | The structure of a transcription activation subcomplex reveals how sigma (70) is recruited to PhoB promoters.
Embo J., 30, 2011
|
|
1DZA
 
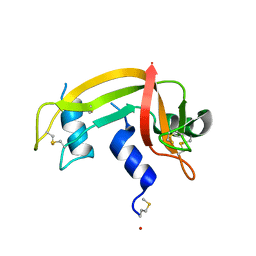 | | 3-D structure of a HP-RNase | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Canals, A, Terzyan, S.S, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-Dimensional Structure of a Human Pancreatic Ribonuclease Variant, a Step Forward in the Design of Cytotoxic Ribonucleases
J.Mol.Biol., 303, 2000
|
|
4JIX
 
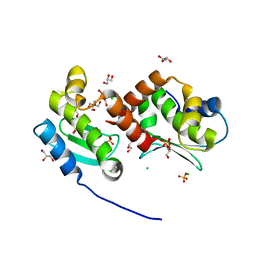 | | Crystal structure of the metallopeptidase zymogen of Methanocaldococcus jannaschii jannalysin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
4JIU
 
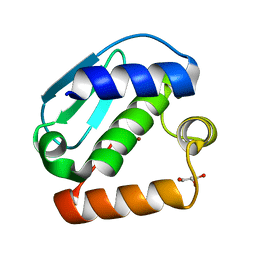 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | Descriptor: | GLYCEROL, Proabylysin, ZINC ION | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
6Y6D
 
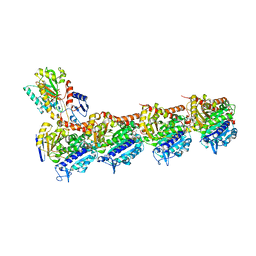 | | Tubulin-7-Aminonoscapine complex | | Descriptor: | (3~{S})-7-azanyl-6-methoxy-3-[(5~{R})-4-methoxy-6-methyl-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3~{H}-2-benzofuran-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Prota, A.E, Rodriguez-Salarichs, J, Gu, W, Bennani, Y.L, Jimenez-Barbero, J, Canales, A, Steinmetz, M.O, Diaz, J.F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Noscapine Activation for Tubulin Binding.
J.Med.Chem., 63, 2020
|
|
2LVZ
 
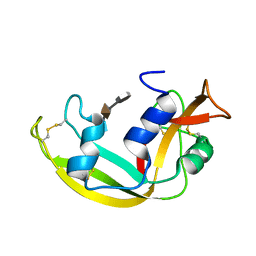 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
4ICV
 
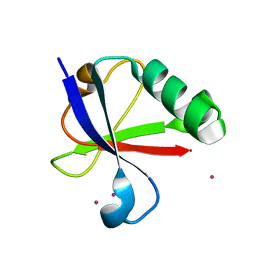 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal form B | | Descriptor: | PRASEODYMIUM ION, Tubulin-specific chaperone E | | Authors: | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | Deposit date: | 2012-12-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
4ICU
 
 | | Ubiquitin-like domain of human tubulin folding cofactor E - crystal from A | | Descriptor: | Tubulin-specific chaperone E | | Authors: | Janowski, R, Boutin, M, Zabala, J.C, Coll, M. | | Deposit date: | 2012-12-11 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the complex between alpha-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism.
J.Cell.Sci., 128, 2015
|
|
6QI4
 
 | | NCS-1 bound to a ligand | | Descriptor: | 2-(1~{H}-indol-3-yl)-~{N}-[(~{E})-(4-nitro-3-oxidanyl-phenyl)methylideneamino]ethanamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Blanco-Gabella, P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into real-time chemical processes in a calcium sensor protein-directed dynamic library.
Nat Commun, 10, 2019
|
|
2N0X
 
 | | Three dimensional structure of EPI-X4, a human albumin-derived peptide that regulates innate immunity through the CXCR4/CXCL12 chemotactic axis and antagonizes HIV-1 entry | | Descriptor: | Serum albumin | | Authors: | Perez-Castells, J, Canales, A, Jimenez-Barbero, J, Gimenez-Gallego, G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and characterization of an endogenous CXCR4 antagonist.
Cell Rep, 11, 2015
|
|
5NJH
 
 | | Triazolopyrimidines stabilize microtubules by binding to the vinca inhibitor site of tubulin | | Descriptor: | 5-chloranyl-7-[(1~{R},5~{S})-3-methoxy-8-azabicyclo[3.2.1]octan-8-yl]-6-[2,4,6-tris(fluoranyl)phenyl]-[1,2,4]triazolo[1,5-a]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Calvo, G.S, Prota, A.E, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Triazolopyrimidines Are Microtubule-Stabilizing Agents that Bind the Vinca Inhibitor Site of Tubulin.
Cell Chem Biol, 24, 2017
|
|
8TJ8
 
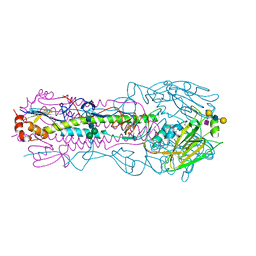 | | CRYSTAL STRUCTURE OF THE A/Moscow/10/1999(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ4
 
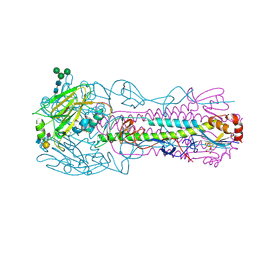 | | CRYSTAL STRUCTURE OF THE A/Bangkok/1/1979(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ9
 
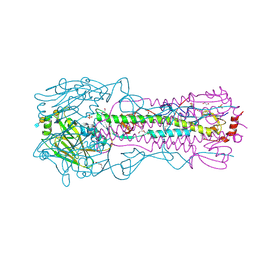 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
