8IRT
 
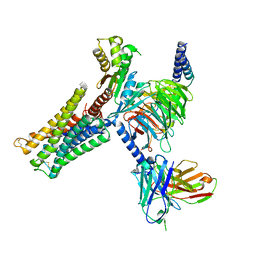 | | Dopamine Receptor D3R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRV
 
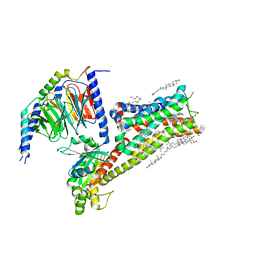 | | Dopamine Receptor D5R-Gs-Rotigotine complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRS
 
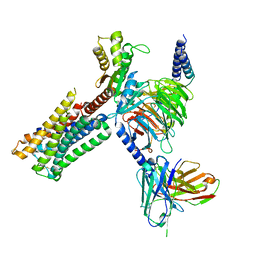 | | Dopamine Receptor D2R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRU
 
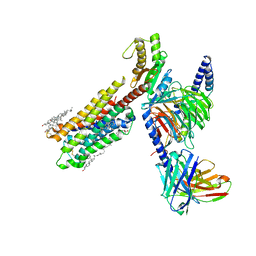 | | Dopamine Receptor D4R-Gi-Rotigotine complex | | Descriptor: | CHOLESTEROL, D(4) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRR
 
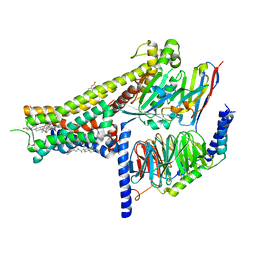 | | Dopamine Receptor D1R-Gs-Rotigotine complex | | Descriptor: | CHOLESTEROL, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
7XZ6
 
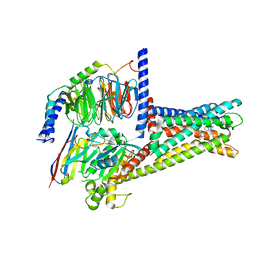 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XZ5
 
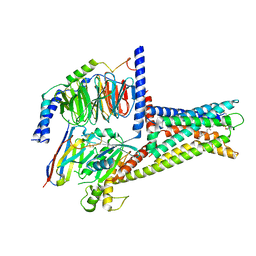 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7CMV
 
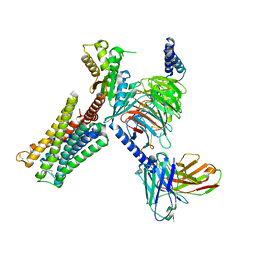 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMU
 
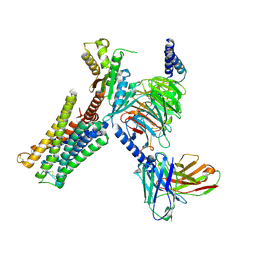 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7E33
 
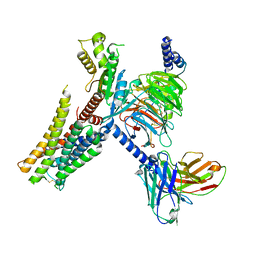 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Y
 
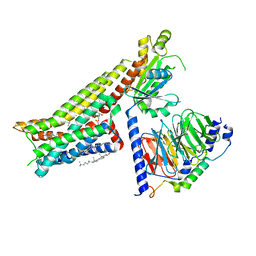 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2X
 
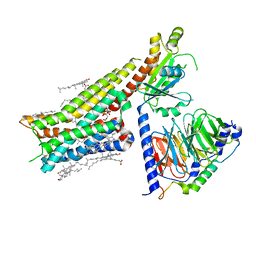 | | Apo serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E32
 
 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
6XJD
 
 | | Two mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Xu, P, Li, P, Zhao, B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
5J9U
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9W
 
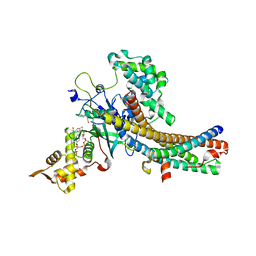 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9T
 
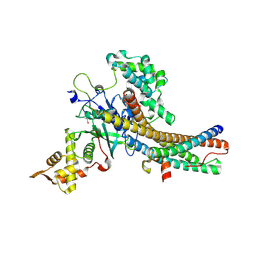 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9Q
 
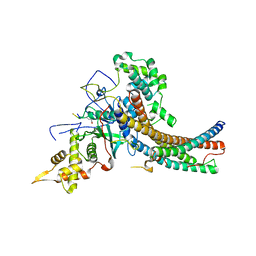 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
7R4H
 
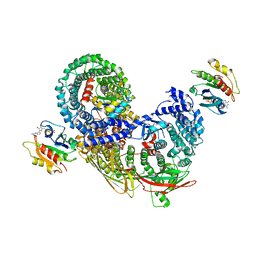 | | phospho-STING binding to adaptor protein complex-1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Xu, P, Ablasser, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Clathrin-associated AP-1 controls termination of STING signalling.
Nature, 610, 2022
|
|
8GKH
 
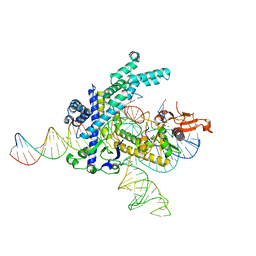 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2023-03-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
7C4A
 
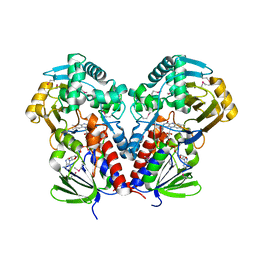 | | nicA2 with cofactor FAD | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7C49
 
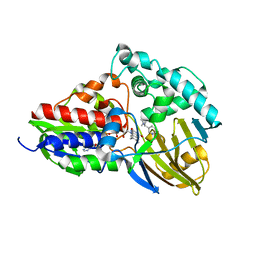 | | nicA2 with cofactor FAD and substrate nicotine | | Descriptor: | 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zhang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
8W89
 
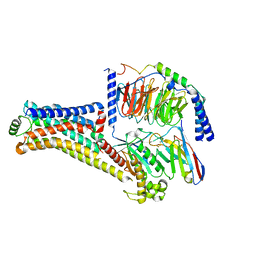 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W87
 
 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
