1A29
 
 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:2 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Bocskei, Zs, Harmat, V, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1998-01-19 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies.
Biochemistry, 37, 1998
|
|
1QIV
 
 | | CALMODULIN COMPLEXED WITH N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE (DPD), 1:2 COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Naray-Szabo, G, Ovadi, J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
1QIW
 
 | | Calmodulin complexed with N-(3,3,-diphenylpropyl)-N'-[1-R-(3,4-bis-butoxyphenyl)-ethyl]-propylenediamine (DPD) | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
3LOJ
 
 | | Structure of Mycobacterium tuberculosis dUTPase H145A mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Pecsi, I, Lopata, A, Vertessy, B.G, Toth, J. | | Deposit date: | 2010-02-04 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase
Nucleic Acids Res., 38, 2010
|
|
3EHW
 
 | | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Takacs, E, Barabas, O, Vertessy, B.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human dUTPase in complex with alpha,beta-imido-dUTP and Mg2+: visualization of the full-length C-termini in all monomers and suggestion for an additional metal ion binding site
To be Published
|
|
2HR6
 
 | | Crystal structure of dUTPase in complex with substrate analogue dUDP and manganese | | Descriptor: | 1,2-ETHANEDIOL, DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
2HRM
 
 | | Crystal structure of dUTPase complexed with substrate analogue methylene-dUTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Tapai, R, Vertessy, B.G. | | Deposit date: | 2006-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site.
Proteins, 71, 2008
|
|
8CGA
 
 | | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, Z.S, Benedek, A, Leveles, I, Vertessy, B.G. | | Deposit date: | 2023-02-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant
To Be Published
|
|
4WRK
 
 | |
4GCY
 
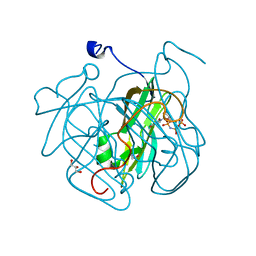 | | Structure of Mycobacterium tuberculosis dUTPase H21W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, J, Vertessy, B.G, Leveles, I, Bendes, A. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RAMD identification of substrate binding pathways to the active site of dUTPase
To be Published
|
|
4GV8
 
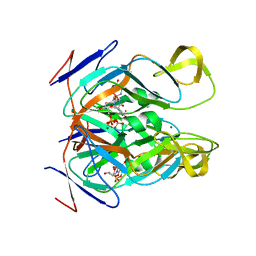 | | DUTPase from phage phi11 of S.aureus: visualization of the species-specific insert | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Leveles, I, Harmat, V, Nemeth, V, Bendes, A, Szabo, J, Kadar, V, Zagyva, I, Rona, G, Toth, J, Vertessy, B.G. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and enzymatic mechanism of a moonlighting dUTPase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8C8I
 
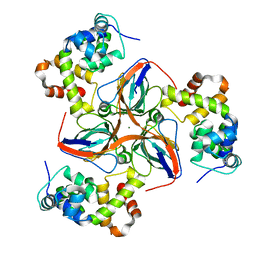 | |
6HDE
 
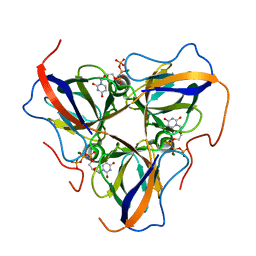 | | Structure of Escherichia coli dUTPase Q93H mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Benedek, A, Vertessy, B.G, Leveles, I. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition.
Biomolecules, 9, 2019
|
|
4MZ6
 
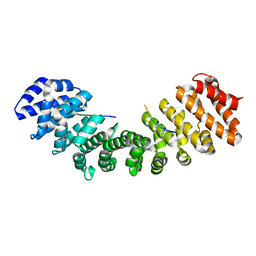 | | Structure of importin-alpha: dUTPase S11E NLS mutant complex | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, Importin subunit alpha-1 | | Authors: | Marfori, M, Rona, G, Vertessy, B.G, Kobe, B. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MZ5
 
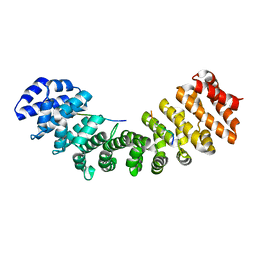 | | Structure of importin-alpha: dUTPase NLS complex | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial, Importin subunit alpha-1 | | Authors: | Marfori, M, Rona, G, Vertessy, B.G, Kobe, B. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylation adjacent to the nuclear localization signal of human dUTPase abolishes nuclear import: structural and mechanistic insights.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2PY4
 
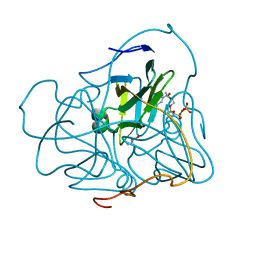 | | Full length structure of the Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Nagy, N, Takacs, E, Vertessy, B.G. | | Deposit date: | 2007-05-15 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor.
Biochem.Biophys.Res.Commun., 373, 2008
|
|
1SYL
 
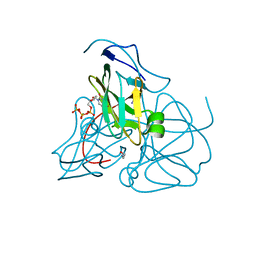 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1RNJ
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RN8
 
 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1SEH
 
 | | Crystal structure of E. coli dUTPase complexed with the product dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
3ECY
 
 | |
2HQU
 
 | | Human dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Varga, B, Vertessy, B.G. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase.
Febs Lett., 581, 2007
|
|
3I93
 
 | | Crystal structure of Mycobacterium tuberculosis dUTPase STOP138T mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-07-10 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
3HZA
 
 | | Crystal structure of dUTPase H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Pecsi, I, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-06-23 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase.
Nucleic Acids Res., 38, 2010
|
|
3IF7
 
 | | Structure of Calmodulin complexed with its first endogenous inhibitor, sphingosylphosphorylcholine | | Descriptor: | 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, CALCIUM ION, Calmodulin | | Authors: | Kovacs, E, Harmat, V, Toth, J, Vertessy, B.G, Modos, K, Kardos, J, Liliom, K. | | Deposit date: | 2009-07-24 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of calmodulin binding to a signaling sphingolipid reveal new aspects of lipid-protein interactions
Faseb J., 24, 2010
|
|
