1P4O
 
 | | Structure of Apo unactivated IGF-1R KInase domain at 1.5A resolution. | | Descriptor: | Insulin-like growth factor I receptor protein | | Authors: | Munshi, S, Kornienko, M, Hall, D.L, Darke, P.L, Waxman, L, Kuo, L.C. | | Deposit date: | 2003-04-23 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1M7N
 
 | |
1C6X
 
 | |
1C6Y
 
 | |
1C70
 
 | |
2BPV
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPY
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPX
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPZ
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | HIV-1 PROTEASE, N-[2(S)-CYCLOPENTYL-1(R)-HYDROXY-3(R)METHYL]-5-[(2(S)-TERTIARY-BUTYLAMINO-CARBONYL)-4-(N1-(2)-(N-METHYLPIPERAZINYL)-3-CHLORO-PYRAZINYL-5-CARBONYL)-PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYL-PENTANAMIDE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2BPW
 
 | | HIV-1 protease-inhibitor complex | | Descriptor: | 1-[2-HYDROXY-4-(2-HYDROXY-5-METHYL-CYCLOPENTYLCARBAMOYL)5-PHENYL-PENTYL]-4-(3-PYRIDIN-3-YL-PROPIONYL)-PIPERAZINE-2-CARB OXYLIC ACID TERT-BUTYLAMIDE, HIV-1 PROTEASE | | Authors: | Munshi, S, Chen, Z. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid X-ray diffraction analysis of HIV-1 protease-inhibitor complexes: inhibitor exchange in single crystals of the bound enzyme.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2IRZ
 
 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | 3-{5-[(1R)-1-AMINO-1-METHYL-2-PHENYLETHYL]-1,3,4-OXADIAZOL-2-YL}-N-[(1R)-1-(4-FLUOROPHENYL)ETHYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
2IS0
 
 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | (2S)-2-AMINO-2-BENZYL-3-HYDROXYPROPYL 3-({[(1R)-1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)-5-[METHYL(METHYLSULFONYL)AMINO]BENZOATE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
2PH6
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | 3-({[(1R)-1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)-5-[METHYL(METHYLSULFONYL)AMINO]BENZYL ALPHA-METHYL-D-PHENYLALANINATE, Beta-secretase 1, SULFATE ION | | Authors: | Munshi, S. | | Deposit date: | 2007-04-10 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QZL
 
 | | Crystal Structure of human Beta Secretase complexed with IXS | | Descriptor: | Beta-secretase 1, N-[(1S)-1-benzyl-2-{[(1S)-2-(isobutylamino)-1-methyl-2-oxoethyl]amino}ethyl]-N'-[(1R)-1-(4-fluorophenyl)ethyl]-5-[methyl(methylsulfonyl)amino]isophthalamide | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BACE-1 inhibition by a series of psi[CH2NH] reduced amide isosteres
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1OHF
 
 | | The refined structure of Nudaurelia capensis omega virus | | Descriptor: | MAGNESIUM ION, p70 | | Authors: | Helgstrand, C, Munshi, S, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Refined Structure of Nudaurelia Capensis Omega Virus Reveals Control Elements for a T = 4 Capsid Maturation
Virology, 318, 2004
|
|
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V2N
 
 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
1CWP
 
 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | Descriptor: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | Authors: | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | Deposit date: | 1995-05-22 | | Release date: | 1995-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
8V5U
 
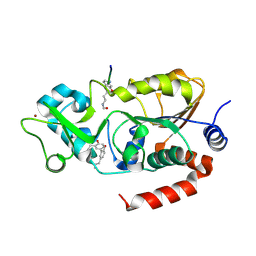 | | Human SIRT3 bound to p53-AMC peptide and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-12-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3-Activating Compounds that Fully Recover Catalytic Activity under ${mathrm{NAD}}^{+}$ Depletion
Phys. Rev. X, 14, 2024
|
|
1C6V
 
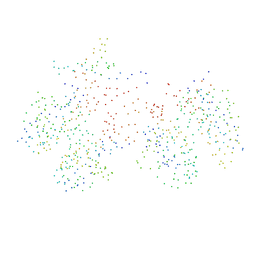 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
1JXP
 
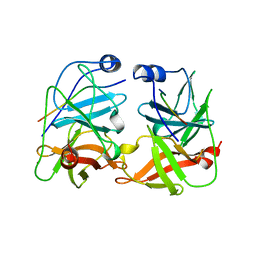 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | Descriptor: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-08-21 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
1NS3
 
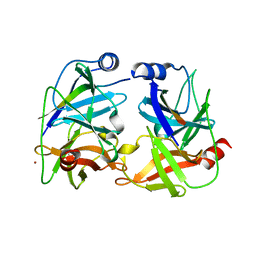 | | STRUCTURE OF HCV PROTEASE (BK STRAIN) | | Descriptor: | NS3 PROTEASE, NS4A PEPTIDE, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
3CUK
 
 | |
