4V8O
 
 | | Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3 | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-07-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Hybrid State of Ribosome in Complex with the Guanosine Triphosphatase Release Factor 3.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V5J
 
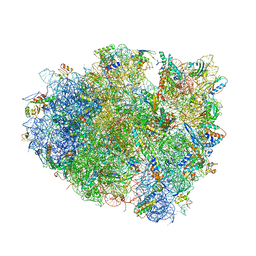 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | Descriptor: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | Deposit date: | 2010-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
3S2Y
 
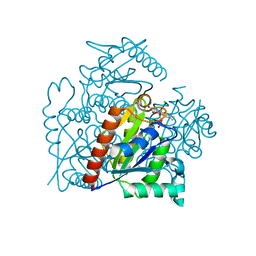 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
2VXW
 
 | |
2PCW
 
 | |
2PCV
 
 | |
5Y7Q
 
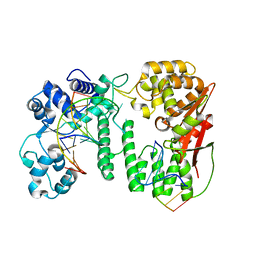 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
5Z6W
 
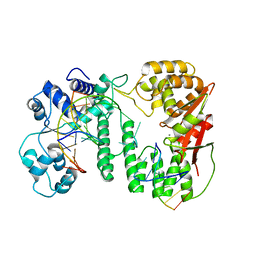 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
6J3Q
 
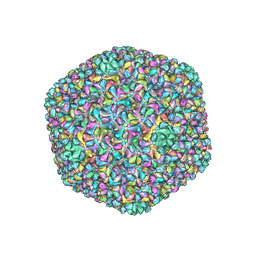 | | Capsid structure of a freshwater cyanophage Siphoviridae Mic1 | | Descriptor: | cement protein, major capsid protein | | Authors: | Jin, H, Jiang, Y.L, Yang, F, Zhang, J.T, Li, W.F, Zhou, K, Ju, J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-05 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Capsid Structure of a Freshwater Cyanophage Siphoviridae Mic1.
Structure, 27, 2019
|
|
1FDP
 
 | | PROENZYME OF HUMAN COMPLEMENT FACTOR D, RECOMBINANT PROFACTOR D | | Descriptor: | PROENZYME OF COMPLEMENT FACTOR D | | Authors: | Jing, H, Macon, K.J, Moore, D, Delucas, L.J, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-12-03 | | Release date: | 1999-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D.
Embo J., 18, 1999
|
|
1BIO
 
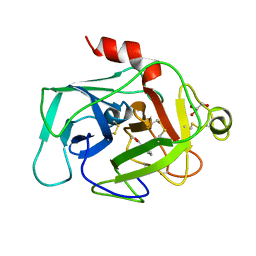 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
1L0Q
 
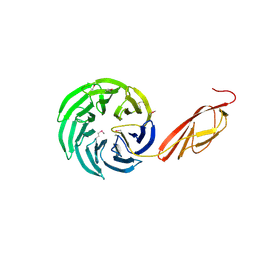 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
1DLE
 
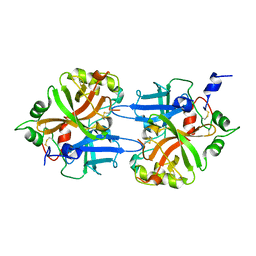 | | FACTOR B SERINE PROTEASE DOMAIN | | Descriptor: | COMPLEMENT FACTOR B | | Authors: | Jing, H, Xu, Y, Carson, M, Moore, D, Macon, K.J, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 1999-12-09 | | Release date: | 2000-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New structural motifs on the chymotrypsin fold and their potential roles in complement factor B.
EMBO J., 19, 2000
|
|
6IHA
 
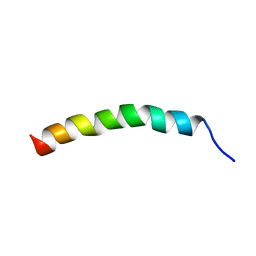 | |
5Y7G
 
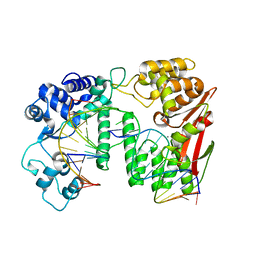 | | Crystal structure of paFAN1 bound to 1nt 5'flap DNA with gap | | Descriptor: | CALCIUM ION, DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Jin, H. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
6JPK
 
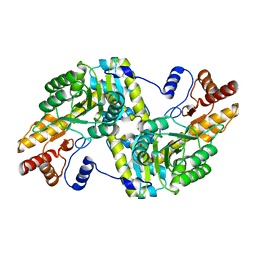 | |
7RR5
 
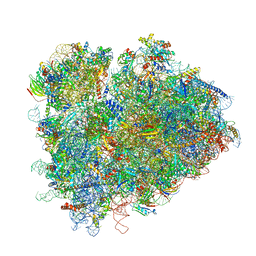 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
9MQ4
 
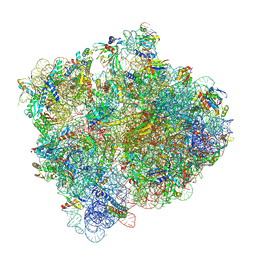 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2025-01-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
9MOR
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2024-12-27 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
4V5E
 
 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
6L2W
 
 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | Descriptor: | freshwater cyanophage protein | | Authors: | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-10-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
5U4I
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5U4J
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
6C4I
 
 | |
