4V5D
 
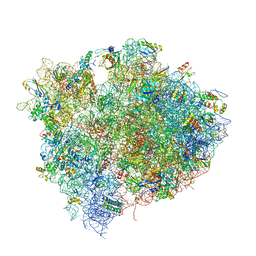 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5C
 
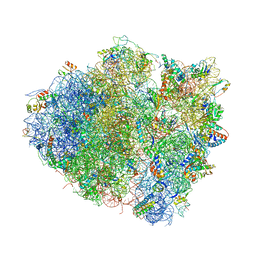 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V5J
 
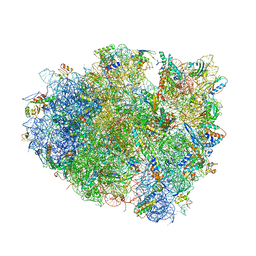 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | Descriptor: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | Deposit date: | 2010-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
417D
 
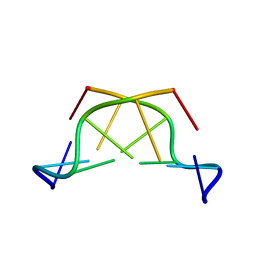 | | A THYMINE-LIKE BASE ANALOGUE FORMS WOBBLE PAIRS WITH ADENINE | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*(C46)P*G)-3') | | Authors: | Lin, P.K.T, Schuerman, M.H, Moore, G.S, Van Meervelt, L, Loakes, D, Brown, D.M, Moore, M.H. | | Deposit date: | 1998-07-15 | | Release date: | 1998-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A thymine-like base analogue forms wobble pairs with adenine in a Z-DNA duplex.
J.Mol.Biol., 282, 1998
|
|
2O4Y
 
 | |
