6AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
5AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
4AT1
 
 | | STRUCTURAL CONSEQUENCES OF EFFECTOR BINDING TO THE T STATE OF ASPARTATE CARBAMOYLTRANSFERASE. CRYSTAL STRUCTURES OF THE UNLIGATED AND ATP-, AND CTP-COMPLEXED ENZYMES AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (T STATE), CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1990-04-26 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution.
Biochemistry, 29, 1990
|
|
2AT2
 
 | |
3BTA
 
 | |
3CZO
 
 | |
2NZ9
 
 | |
2NYY
 
 | |
2PAH
 
 | |
1ACM
 
 | | ARGININE 54 IN THE ACTIVE SITE OF ESCHERICHIA COLI ASPARTATE TRANSCARBAMOYLASE IS CRITICAL FOR CATALYSIS: A SITE-SPECIFIC MUTAGENESIS, NMR AND X-RAY CRYSTALLOGRAPHY STUDY | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN, ... | | Authors: | Stevens, R.C, Kantrowitz, E.R, Lipscomb, W.N. | | Deposit date: | 1992-07-08 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study.
Protein Sci., 1, 1992
|
|
1HTZ
 
 | | CRYSTAL STRUCTURE OF TEM52 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE MUTANT TEM52 | | Authors: | Stevens, R.C, Orencia, M.C. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predicting the emergence of antibiotic resistance by directed evolution and structural analysis.
Nat.Struct.Biol., 8, 2001
|
|
1FL3
 
 | |
1PAH
 
 | |
8AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
7AT1
 
 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
5XR8
 
 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
3K84
 
 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
3K3Q
 
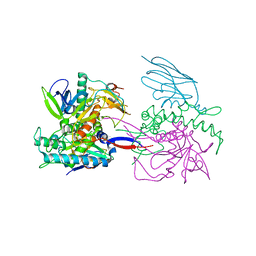 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
5DHH
 
 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with SB-612111 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5S,7S)-7-{[4-(2,6-dichlorophenyl)piperidin-1-yl]methyl}-1-methyl-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ol, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
2VI6
 
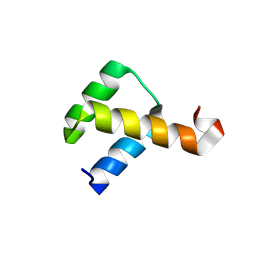 | | Crystal Structure of the Nanog Homeodomain | | Descriptor: | HOMEOBOX PROTEIN NANOG | | Authors: | Jauch, R, Ng, C.K.L, Saitakendu, K.S, Stevens, R.C, Kolatkar, P.R. | | Deposit date: | 2007-11-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog.
J.Mol.Biol., 376, 2008
|
|
4L6R
 
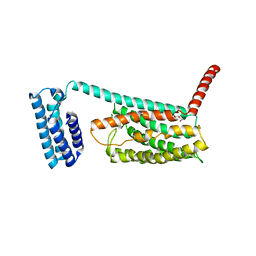 | | Structure of the class B human glucagon G protein coupled receptor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Soluble cytochrome b562 and Glucagon receptor chimera | | Authors: | Siu, F.Y, He, M, de Graaf, C, Han, G.W, Yang, D, Zhang, Z, Zhou, C, Xu, Q, Wacker, D, Joseph, J.S, Liu, W, Lau, J, Cherezov, V, Katritch, V, Wang, M.W, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the human glucagon class B G-protein-coupled receptor.
Nature, 499, 2013
|
|
7DTY
 
 | | Structural basis of ligand selectivity conferred by the human glucose-dependent insulinotropic polypeptide receptor | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhang, C, Zhou, Q.T, Hang, K.N, Zou, X.Y, Chen, Y, Wu, F, Rao, Q.D, Dai, A.T, Yin, W.C, Shen, D.D, Zhang, Y, Xia, T, Stevens, R.C, Xu, H.E, Yang, D.H, Zhao, L.H, Wang, M.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Elife, 10, 2021
|
|
6W25
 
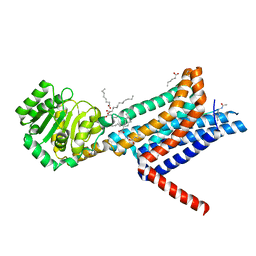 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
