5AX1
 
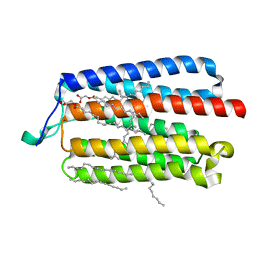 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5OUJ
 
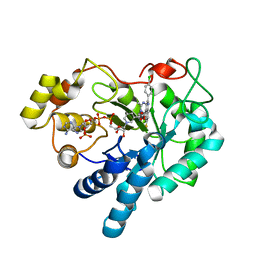 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OUK
 
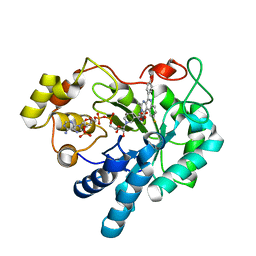 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OU0
 
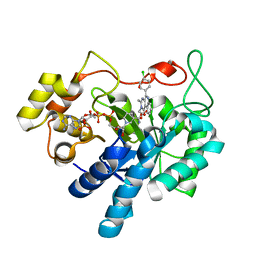 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
5UDH
 
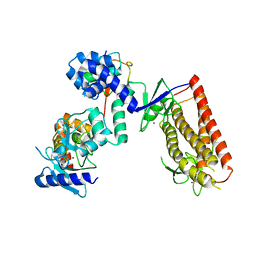 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5TTE
 
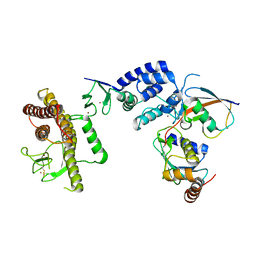 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
2ZJ1
 
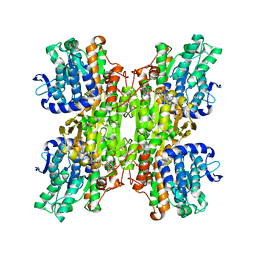 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 3'-keto-aristeromycin | | Descriptor: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
2ZJ0
 
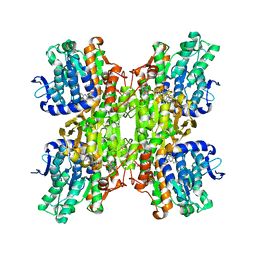 | | Crystal structure of Mycobacterium tuberculosis S-Adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
2ZIZ
 
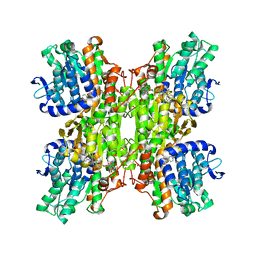 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 3-deazaadenosine | | Descriptor: | 3-DEAZA-ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
3CE6
 
 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
7B5N
 
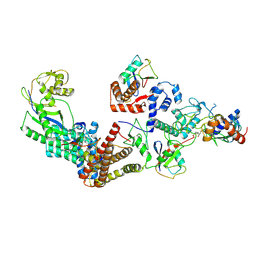 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5L
 
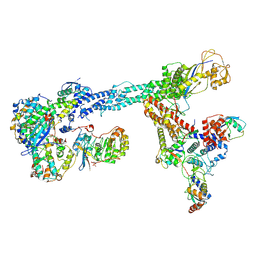 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, Cyclin-A2, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5S
 
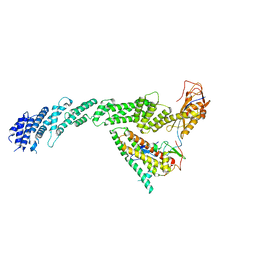 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5M
 
 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
4GAB
 
 | | Human AKR1B10 mutant V301L complexed with NADP+ and fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldo-keto reductase family 1 member B10, CHLORIDE ION, ... | | Authors: | Cousido-Siah, A, Ruiz Figueras, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2012-07-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5971 Å) | | Cite: | X-ray structure of the V301L aldo-keto reductase 1B10 complexed with NADP(+) and the potent aldose reductase inhibitor fidarestat: Implications for inhibitor binding and selectivity.
Chem.Biol.Interact, 202, 2013
|
|
1M25
 
 | | STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE IN TFE SOLUTION | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Megy, S, Bertho, G, Kozin, S.A, Coadou, G, Debey, P, Hoa, G.H, Girault, J.-P. | | Deposit date: | 2002-06-21 | | Release date: | 2002-07-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Possible role of region 152-156 in the structural duality of a peptide fragment from sheep prion protein
Protein Sci., 13, 2004
|
|
2BTY
 
 | | Acetylglutamate kinase from Thermotoga maritima complexed with its inhibitor arginine | | Descriptor: | ACETYLGLUTAMATE KINASE, ARGININE, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Gil-Ortiz, F, Fernandez-Murga, M.L, Fita, I, Rubio, V. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Bases of Feed-Back Control of Arginine Biosynthesis, Revealed by the Structure of Two Hexameric N-Acetylglutamate Kinases, from Thermotoga Maritima and Pseudomonas Aeruginosa
J.Mol.Biol., 356, 2006
|
|
3DHY
 
 | | Crystal Structures of Mycobacterium tuberculosis S-Adenosyl-L-Homocysteine Hydrolase in Ternary Complex with Substrate and Inhibitors | | Descriptor: | 5'-S-ethyl-5'-thioadenosine, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shetty, N.D, Ioerger, T.R, Gokulan, K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-06-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
4KBL
 
 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
4KC9
 
 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
2M9Y
 
 | |
7OD1
 
 | |
7ONI
 
 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
8Q5R
 
 | | HpARI bound to mouse IL-33 | | Descriptor: | 1,2-ETHANEDIOL, Alarmin release inhibitor, CHLORIDE ION, ... | | Authors: | Jamwal, A, Higgins, M.K. | | Deposit date: | 2023-08-09 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for IL-33 recognition and its antagonism by the helminth effector protein HpARI2
To Be Published
|
|
