5JIX
 
 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
5JIZ
 
 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP
To Be Published
|
|
4D7S
 
 | | Structure of the SthK Carboxy-Terminal Region in complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, STHK_CNBD_CGMP | | Authors: | Kesters, D, Brams, M, Nys, M, Wijckmans, E, Spurny, R, Voets, T, Tytgat, J, Ulens, C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels.
Plos One, 10, 2015
|
|
4D7T
 
 | | Structure of the SthK Carboxy-Terminal Region in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, STHK_CNBD_CAMP | | Authors: | Kesters, D, Brams, M, Nys, M, Wijckmans, E, Spurny, R, Voets, T, Tytgat, J, Ulens, C. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Structure of the SthK Carboxy-Terminal Region Reveals a Gating Mechanism for Cyclic Nucleotide-Modulated Ion Channels.
Plos One, 10, 2015
|
|
5KBF
 
 | | cAMP bound PfPKA-R (141-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit, putative | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
7LZ4
 
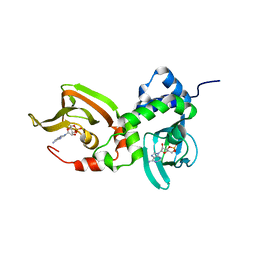 | | Crystal structure of A211D mutant of Protein Kinase A RIa subunit, a Carney Complex mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit, N-terminally processed | | Authors: | Del Rio, J, Wu, J, Taylor, S.S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.155 Å) | | Cite: | Noncanonical protein kinase A activation by oligomerization of regulatory subunits as revealed by inherited Carney complex mutations.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6FTF
 
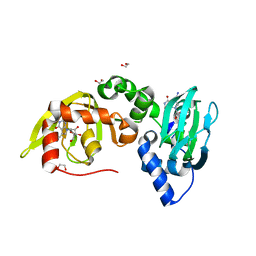 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.09 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-cyano-7-deazainosine, Protein kinase A regulatory subunit, ... | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09151936 Å) | | Cite: | Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma.
Nat Commun, 10, 2019
|
|
6FLO
 
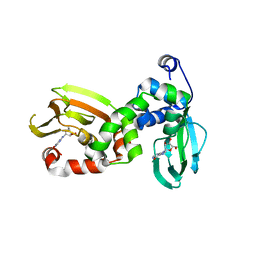 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei at 2.1 Angstrom resolution | | Descriptor: | GLYCEROL, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-01-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.13868666 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
6H4G
 
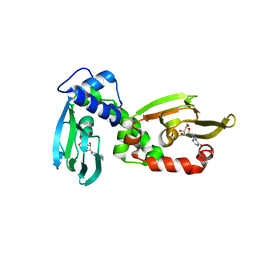 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma brucei: E311A, T318R, V319A mutant bound to cAMP in the A site | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, INOSINE, Protein kinase A regulatory subunit | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-07-21 | | Release date: | 2019-08-14 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.143517 Å) | | Cite: | Purine nucleosides replace cAMP in allosteric regulation of PKA in trypanosomatid pathogens.
Elife, 12, 2024
|
|
6HQ3
 
 | |
4F8A
 
 | | Cyclic nucleotide binding-homology domain from mouse EAG1 potassium channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 1 | | Authors: | Marques-Carvalho, M.J, Sahoo, N, Muskett, F.W, Vieira-Pires, R.S, Gabant, G, Cadene, M, Schonherr, R, Morais-Cabral, J.H. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Characterization of the Cyclic Nucleotide Binding Homology Domain from the Mouse EAG1 Potassium Channel.
J.Mol.Biol., 423, 2012
|
|
5BV6
 
 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMP | | Descriptor: | ACETATE ION, CALCIUM ION, GUANOSINE-3',5'-MONOPHOSPHATE, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5C6C
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5C8W
 
 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MALONIC ACID, SODIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
4HBN
 
 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
5D1I
 
 | |
5E16
 
 | | Co-crystal structure of the N-termial cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4JVA
 
 | | Crystal Structure of RIIbeta(108-402) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
4JV4
 
 | | Crystal Structure of RIalpha(91-379) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
1NE6
 
 | | Crystal structure of Sp-cAMP binding R1a subunit of cAMP-dependent protein kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
1NE4
 
 | | Crystal Structure of Rp-cAMP Binding R1a Subunit of cAMP-dependent Protein Kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
1Q5O
 
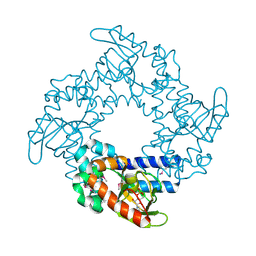 | | HCN2J 443-645 in the presence of cAMP, selenomethionine derivative | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STRUCTURAL BASIS FOR MODULATION AND AGONIST SPECIFICITY OF HCN PACEMAKER CHANNELS
Nature, 425, 2003
|
|
1Q43
 
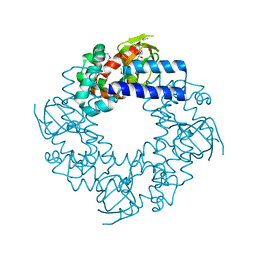 | | HCN2I 443-640 in the presence of cAMP, selenomethionine derivative | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-08-01 | | Release date: | 2003-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
1Q3E
 
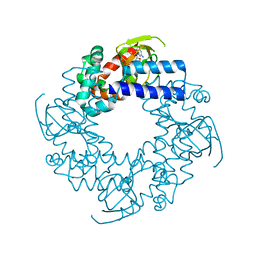 | | HCN2J 443-645 in the presence of cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
1RL3
 
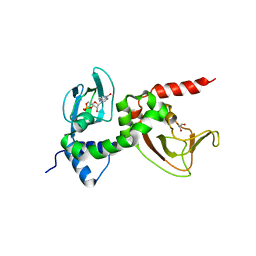 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
