1SNB
 
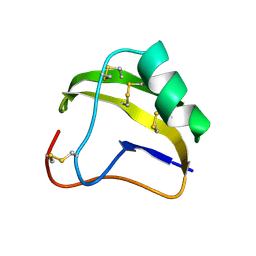 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1SNK
 
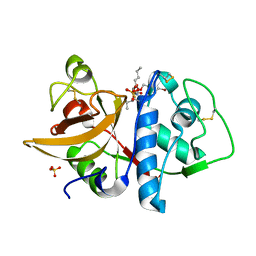 | | Cathepsin K complexed with carbamate derivatized norleucine aldehyde | | Descriptor: | Cathepsin K, N2-({[(4-BROMOPHENYL)METHYL]OXY}CARBONYL)-N1-[(1S)-1-FORMYLPENTYL]-L-LEUCINAMIDE, SULFATE ION | | Authors: | Boros, E.E, Deaton, D.N, Hassell, A.M, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M, Thompson, J.B, Willard Jr, D.H, Wright, L.L. | | Deposit date: | 2004-03-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploration of the P(2)-P(3) SAR of aldehyde cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S8D
 
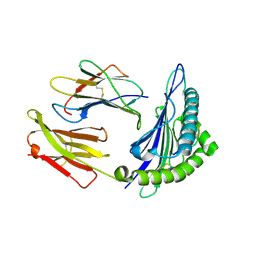 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-3A | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-02-02 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1SCI
 
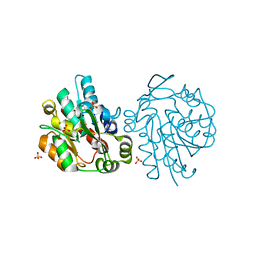 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | Descriptor: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SPP
 
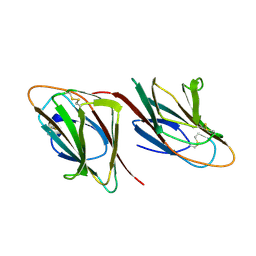 | | THE CRYSTAL STRUCTURES OF TWO MEMBERS OF THE SPERMADHESIN FAMILY REVEAL THE FOLDING OF THE CUB DOMAIN | | Descriptor: | MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-I, MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-II | | Authors: | Romero, A, Romao, M.J, Varela, P.F, Kolln, I, Dias, J.M, Carvalho, A.L, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-19 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
1S9Q
 
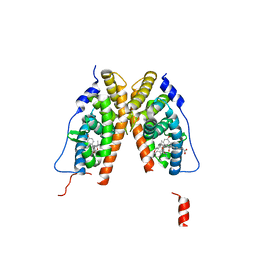 | | crystal structure of the ligand-binding domain of the estrogen-related receptor gamma in complex with 4-hydroxytamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, CHOLIC ACID, Estrogen-related receptor gamma | | Authors: | Greschik, H, Flaig, R, Renaud, J.P, Moras, D. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Deactivation of the Estrogen-related Receptor {gamma} by Diethylstilbestrol or 4-Hydroxytamoxifen and Determinants of Selectivity.
J.Biol.Chem., 279, 2004
|
|
1SBB
 
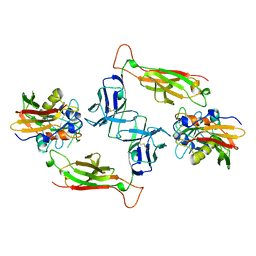 | | T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SUPERANTIGEN SEB | | Descriptor: | PROTEIN (14.3.D T CELL ANTIGEN RECEPTOR), PROTEIN (STAPHYLOCOCCAL ENTEROTOXIN B) | | Authors: | Li, H, Mariuzza, R.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B.
Immunity, 9, 1998
|
|
1SKS
 
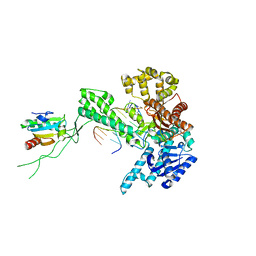 | | Binary 3' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template | | Descriptor: | 5'-D(*CP*CP*CP*(TTD)P*AP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*C*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*(2DT))-3', DNA polymerase, ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SC8
 
 | | Urokinase Plasminogen Activator B-Chain-J435 Complex | | Descriptor: | N-(BENZYLSULFONYL)SERYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}GLYCINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
1SCK
 
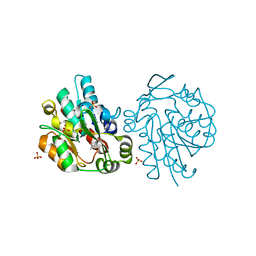 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | Descriptor: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SDR
 
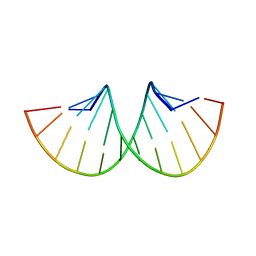 | | CRYSTAL STRUCTURE OF AN RNA DODECAMER CONTAINING THE ESCHERICHIA COLI SHINE-DALGARNO SEQUENCE | | Descriptor: | RNA (5'-R(*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3'), RNA (5'-R(*UP*AP*AP*GP*GP*AP*GP*GP*UP*GP*AP*U)-3') | | Authors: | Schindelin, H, Zhang, M, Bald, R, Fuerste, J.-P, Erdmann, V.A, Heinemann, U. | | Deposit date: | 1994-12-11 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence.
J.Mol.Biol., 249, 1995
|
|
1SI4
 
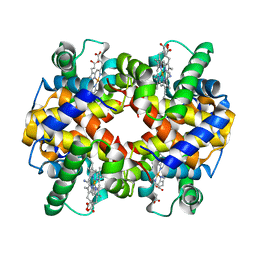 | | Crystal structure of Human hemoglobin A2 (in R2 state) at 2.2 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin delta chain, ... | | Authors: | Sen, U, Dasgupta, J, Choudhury, D, Datta, P, Chakrabarti, A, Chakrabarty, S.B, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2004-02-27 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of HbA2 and HbE and modeling of hemoglobin delta4: interpretation of the thermal stability and the antisickling effect of HbA2 and identification of the ferrocyanide binding site in Hb.
Biochemistry, 43, 2004
|
|
1SEB
 
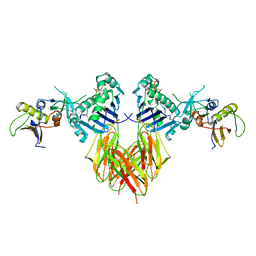 | | COMPLEX OF THE HUMAN MHC CLASS II GLYCOPROTEIN HLA-DR1 AND THE BACTERIAL SUPERANTIGEN SEB | | Descriptor: | ENDOGENOUS PEPTIDE MODEL, POLY-ALA, ENTEROTOXIN TYPE B, ... | | Authors: | Jardetzky, T.S, Brown, J.H, Gorga, J.C, Stern, L.J, Urban, R.G, Chi, Y.I, Stauffacher, C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1995-11-26 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen.
Nature, 368, 1994
|
|
1SIV
 
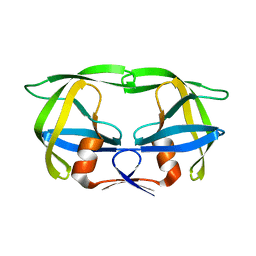 | |
1SMF
 
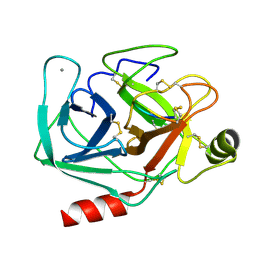 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
1SJF
 
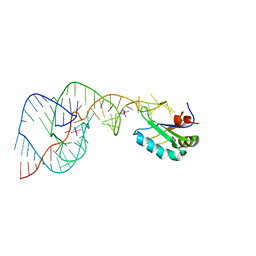 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Cobalt Hexammine solution | | Descriptor: | COBALT HEXAMMINE(III), Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A conformational switch controls hepatitis delta virus ribozyme catalysis.
Nature, 429, 2004
|
|
1SN4
 
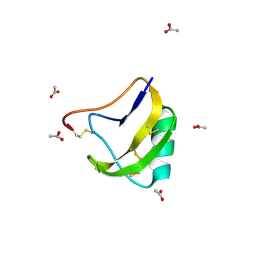 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SL1
 
 | | Binary 5' complex of T7 DNA polymerase with a DNA primer/template containing a cis-syn thymine dimer on the template | | Descriptor: | 5'-D(*CP*CP*C*(TTD)P*AP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*TP*(2DA))-3', DNA polymerase, ... | | Authors: | Li, Y, Dutta, S, Doublie, S, Bdour, H.M, Taylor, J.S, Ellenberger, T. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SMN
 
 | |
1SCF
 
 | | HUMAN RECOMBINANT STEM CELL FACTOR | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, STEM CELL FACTOR | | Authors: | Jiang, X, Gurel, O, Langley, K.E, Hendrickson, W.A. | | Deposit date: | 1998-06-04 | | Release date: | 2000-07-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the active core of human stem cell factor and analysis of binding to its receptor kit.
EMBO J., 19, 2000
|
|
1SPJ
 
 | | STRUCTURE OF MATURE HUMAN TISSUE KALLIKREIN (HUMAN KALLIKREIN 1 OR KLK1) AT 1.70 ANGSTROM RESOLUTION WITH VACANT ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Laxmikanthan, G, Blaber, S.I, Bernett, M.J, Blaber, M. | | Deposit date: | 2004-03-16 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 A X-ray structure of human apo kallikrein 1: Structural changes upon peptide inhibitor/substrate binding
Proteins, 58, 2005
|
|
1SCM
 
 | |
1R9H
 
 | |
1RDN
 
 | | MANNOSE-BINDING PROTEIN, SUBTILISIN DIGEST FRAGMENT COMPLEX WITH ALPHA-METHYL-D-N-ACETYLGLUCOSAMINIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ng, K.K.-S, Drickamer, K, Weis, W.I. | | Deposit date: | 1995-09-05 | | Release date: | 1996-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of monosaccharide recognition by rat liver mannose-binding protein.
J.Biol.Chem., 271, 1996
|
|
1RE1
 
 | |
