2R5M
 
 | |
2V5X
 
 | | Crystal structure of HDAC8-inhibitor complex | | Descriptor: | (2R)-N~8~-HYDROXY-2-{[(5-METHOXY-2-METHYL-1H-INDOL-3-YL)ACETYL]AMINO}-N~1~-[2-(2-PHENYL-1H-INDOL-3-YL)ETHYL]OCTANEDIAMIDE, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C, Gallinari, P, Jones, P, Mattu, M, Carfi, A, Defrancesco, R, Steinkuhler, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
2VI6
 
 | | Crystal Structure of the Nanog Homeodomain | | Descriptor: | HOMEOBOX PROTEIN NANOG | | Authors: | Jauch, R, Ng, C.K.L, Saitakendu, K.S, Stevens, R.C, Kolatkar, P.R. | | Deposit date: | 2007-11-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and DNA Binding of the Homeodomain of the Stem Cell Transcription Factor Nanog.
J.Mol.Biol., 376, 2008
|
|
2VL2
 
 | | Oxidized and reduced forms of human peroxiredoxin 5 | | Descriptor: | BENZOIC ACID, PEROXIREDOXIN-5 | | Authors: | Smeets, A, Declercq, J.P. | | Deposit date: | 2008-01-08 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The Crystal Structures of Oxidized Forms of Human Peroxiredoxin 5 with an Intramolecular Disulfide Bond Confirm the Proposed Enzymatic Mechanism for Atypical 2-Cys Peroxiredoxins.
Arch.Biochem.Biophys., 477, 2008
|
|
4MLP
 
 | |
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3H7H
 
 | | Crystal structure of the human transcription elongation factor DSIF, hSpt4/hSpt5 (176-273) | | Descriptor: | BETA-MERCAPTOETHANOL, Transcription elongation factor SPT4, Transcription elongation factor SPT5, ... | | Authors: | Wenzel, S, Wohrl, B.M, Rosch, P, Martins, B.M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface
Biochem.J., 425, 2010
|
|
3C5H
 
 | | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP) | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP).
To be Published
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
3BIP
 
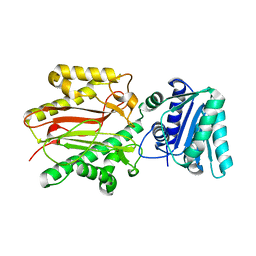 | | Crystal structure of yeast Spt16 N-terminal Domain | | Descriptor: | FACT complex subunit SPT16 | | Authors: | VanDemark, A.P, Xin, H, McCullough, L, Rawlins, R, Bentley, S, Heroux, A, David, S.J, Hill, C.P, Formosa, T. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits.
J.Biol.Chem., 283, 2008
|
|
3B6Y
 
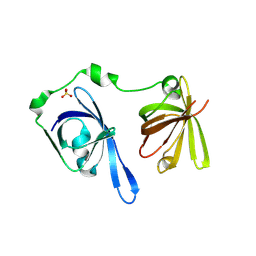 | | Crystal Structure of the Second HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | Gamma-interferon-inducible protein Ifi-16, SULFATE ION | | Authors: | Liao, J.C.C, Lam, R, Ravichandran, M, Duan, S, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure Analysis of the Second HIN Domain of IFI16.
To be Published
|
|
7YI5
 
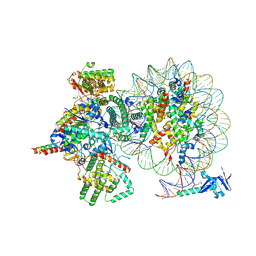 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
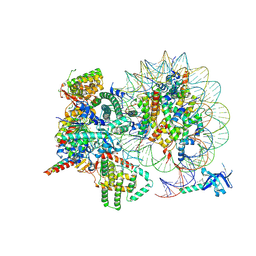 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7Y8R
 
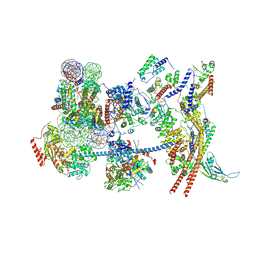 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
4OJJ
 
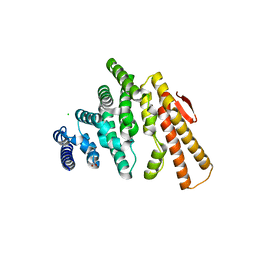 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4PX8
 
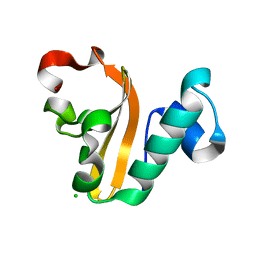 | | Structure of P. vulgaris HigB toxin | | Descriptor: | CHLORIDE ION, Killer protein | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2014-03-22 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3B5B
 
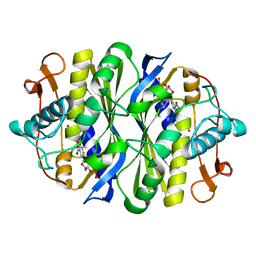 | | Crystal structure of the thymidylate synthase k48q | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-(DIHYDROGEN PHOSPHATE), 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, FORMIC ACID, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of an invariant lysine residue in folate binding on Escherichia coli thymidylate synthase: Calorimetric and crystallographic analysis of the K48Q mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
4OM2
 
 | |
4OP0
 
 | | Crystal structure of biotin protein ligase (RV3279C) of Mycobacterium tuberculosis, complexed with biotinyl-5'-AMP | | Descriptor: | BIOTINYL-5-AMP, BirA bifunctional protein, SULFATE ION | | Authors: | Ma, Q, Wilmanns, M, Akhter, Y. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active site conformational changes upon reaction intermediate biotinyl-5'-AMP binding in biotin protein ligase from Mycobacterium tuberculosis.
Protein Sci., 23, 2014
|
|
3DXB
 
 | | Structure of the UHM domain of Puf60 fused to thioredoxin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, thioredoxin N-terminally fused to Puf60(UHM) | | Authors: | Corsini, L, Hothorn, M, Scheffzek, K, Stier, G, Sattler, M. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimerization and Protein Binding Specificity of the U2AF Homology Motif of the Splicing Factor Puf60.
J.Biol.Chem., 284, 2009
|
|
4QHV
 
 | | Crystal structure of human dihydrofolate reductase as complex with pyridopyrimidine 22 (N~6~-METHYL-N~6~-[4-(PROPAN-2-YL)PHENYL]PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-[4-(propan-2-yl)phenyl]pyrido[2,3-d]pyrimidine-2,4,6-triamine, ... | | Authors: | Cody, V. | | Deposit date: | 2014-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4QJC
 
 | | Human dihydrofolate reductase ternary complex with NADPH and inhibitor 26 (N~6~-METHYL-N~6~-(3,4,5-TRIFLUOROPHENYL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(3,4,5-trifluorophenyl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2014-06-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3EIG
 
 | |
3TPE
 
 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
