7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
8HAF
 
 | | PTHrP-PTH1R-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | Descriptor: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8QQK
 
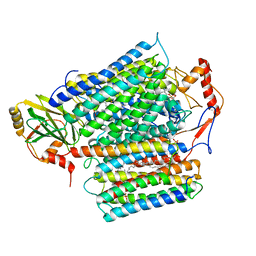 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
1FR8
 
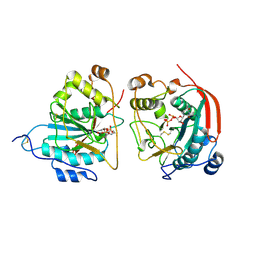 | | CRYSTAL STRUCTURE OF THE BOVINE BETA 1,4 GALACTOSYLTRANSFERASE (B4GALT1) CATALYTIC DOMAIN COMPLEXED WITH URIDINE DIPHOSPHOGALACTOSE | | Descriptor: | BETA 1,4 GALACTOSYLTRANSFERASE, GALACTOSE-URIDINE-5'-DIPHOSPHATE | | Authors: | Gastinel, L.N, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-07 | | Release date: | 2000-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose.
EMBO J., 18, 1999
|
|
8ELF
 
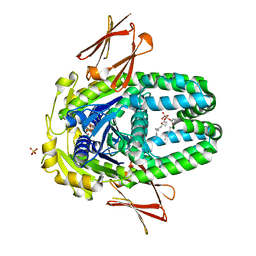 | | Structure of Get3d, a homolog of Get3, from Arabidopsis thaliana | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Manu, M.S, Barlow, A.N, Clemons Jr, W.M, Ramasamy, S. | | Deposit date: | 2022-09-23 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
5OQ3
 
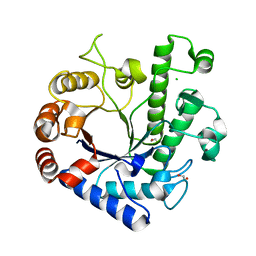 | | High resolution structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cwp19, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
7X8O
 
 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
4UAL
 
 | | MRCK beta in complex with BDP00005290 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-1-(piperidin-4-yl)-N-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
1FHW
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
8KEE
 
 | | Cyanophage A-1(L) sheath-tube | | Descriptor: | sheath, tube | | Authors: | Yu, R.C, Li, Q, Zhou, C.Z. | | Deposit date: | 2023-08-11 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the intact tail machine of Anabaena myophage A-1(L).
Nat Commun, 15, 2024
|
|
8QUC
 
 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1 | | Descriptor: | (5R)-5-ethyl-3-[6-(3-methoxy-4-methyl-phenoxy)pyridin-3-yl]imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G, Large, C, Lakshminaraya, B, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
1FKX
 
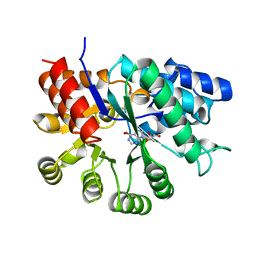 | | MURINE ADENOSINE DEAMINASE (D296A) | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
9BY6
 
 | |
6ZUJ
 
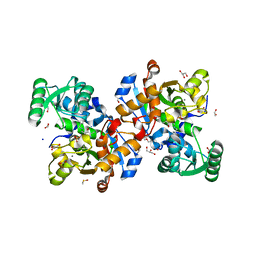 | | Human serine racemase holoenzyme from 20% DMSO soak (XChem crystallographic fragment screen). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
5OOU
 
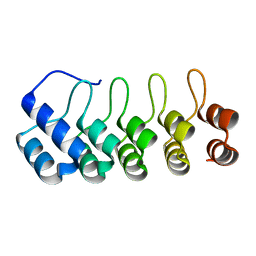 | | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme | | Descriptor: | DARPin YTRL-1 | | Authors: | Fischer, G, Hogan, B.J, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) YTRL-1 selected by directed evolution against Lysozyme
To be published
|
|
8VZK
 
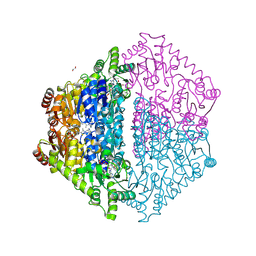 | | Crystal Structure of 2-Hydroxyacyl-CoA Lyase/Synthase CfhHACS from Chloroflexi bacterium in the Complex with THDP and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Gade, P, Mayfield, A, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2024-02-11 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8QP8
 
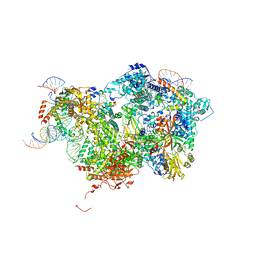 | | Cryo-EM Structure of Pre-B Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
7R9L
 
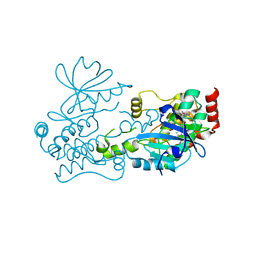 | | Crystal structure of HPK1 in complex with compound 2 | | Descriptor: | 2-amino-N,N-dimethyl-5-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
4UST
 
 | |
8KEF
 
 | |
8KEG
 
 | |
7OGY
 
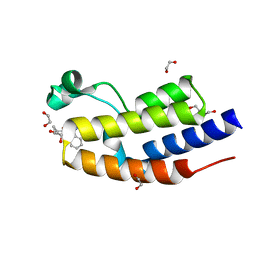 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1-benzyl-N5-cyclopropyl-N3-methyl-1H-pyrazole-3,5-dicarboxamide | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Scaffold Hopping: Identification of Potent, Selective, and Highly Soluble Bromo and Extra Terminal Domain (BET) Second Bromodomain (BD2) Inhibitors.
J.Med.Chem., 64, 2021
|
|
5O9E
 
 | | Crystal structure of the Imp4-Mpp10 complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, Putative U3 small nucleolar ribonucleoprotein, Putative U3 small nucleolar ribonucleoprotein protein | | Authors: | Kharde, S, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2017-06-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Mpp10 represents a platform for the interaction of multiple factors within the 90S pre-ribosome.
PLoS ONE, 12, 2017
|
|
