6Z6L
 
 | | Cryo-EM structure of human CCDC124 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
5GAG
 
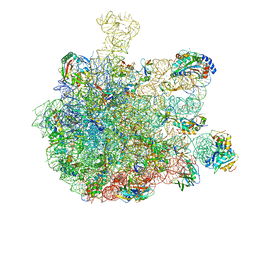 | | RNC in complex with SRP-SR in the closed state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
5KMZ
 
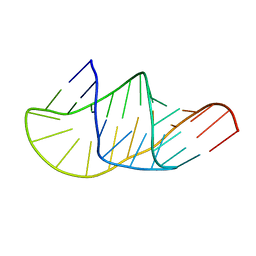 | |
9G8N
 
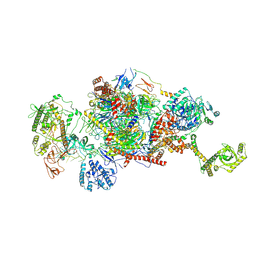 | | 80S-bound human Ski2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 2024
|
|
9G8P
 
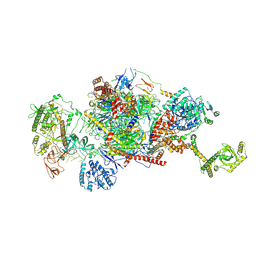 | | 40S-bound human SKI2-exosome complex | | Descriptor: | CrPV-IRES RNA, DIS3-like exonuclease 1, Exosome complex component CSL4, ... | | Authors: | Koegel, A, Keidel, A, Loukeri, M.J, Kuhn, C.C, Langer, L.M, Schaefer, I.B, Conti, E. | | Deposit date: | 2024-07-23 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of mRNA decay by the human exosome-ribosome supercomplex.
Nature, 2024
|
|
5G4N
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a difluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | 1-[9-(2,2-difluoroethyl)-9H-carbazol-3-yl]-N-methylmethanamine, CELLULAR TUMOR ANTIGEN P53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M, Jones, R.N, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5G4M
 
 | |
5FA5
 
 | |
5G4O
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a trifluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, N,N-dimethyl-1-[9-(2,2,2-trifluoroethyl)-9H-carbazol-3-yl]methanamine, ZINC ION | | Authors: | Joerger, A.C, Bauer, M, Baud, M.G.J, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
6VRD
 
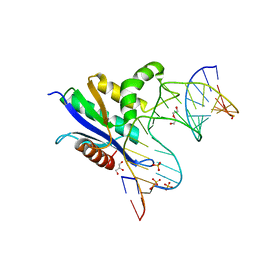 | | Crystal structure of RNase H/RNA/PS-ASO complex at an atomic level | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RNA (5'-R(*(OMC)P*(N7X)P*(T39)P*(C5L)P*(A2M))-D(P*(SC)P*(PST)P*(SC)P*(AS)P*(SC)P*(SC)P*(SC)P*(AS)P*(SC)P*(PST))-R(P*(6OO)P*(RFJ)P*(6OO)P*(6OO)P*(6NW))-3'), ... | | Authors: | Cho, Y.-J, Butler, D. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Crystal structure of RNase H/RNA/PS-ASO complex at an atomic level
To Be Published
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2BL7
 
 | | 1.6 Angstrom crystal structure of EntA-im: a bacterial immunity protein conferring immunity to the antimicrobial activity of the pediocin-like bacteriocin, enterocin A | | Descriptor: | ENTEROCINE A IMMUNITY PROTEIN | | Authors: | Johnsen, L, Dalhus, B, Leiros, I, Nissen-Meyer, J. | | Deposit date: | 2005-03-02 | | Release date: | 2005-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 1.6-A Crystal Structure of Enta-Im: A Bacterial Immunity Protein Conferring Immunity to the Antimicrobial Activity of the Pediocin-Like Bacteriocin Enterocin A
J.Biol.Chem., 280, 2005
|
|
6VXU
 
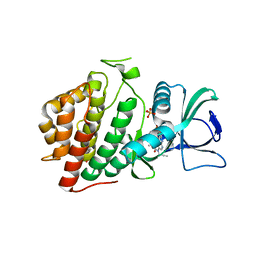 | | Structure of Human Vaccinia-related Kinase 1 (VRK1) bound to ACH471 | | Descriptor: | (7R)-8-(cyclopropylmethyl)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, (7S)-8-(cyclopropylmethyl)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, GLYCEROL, ... | | Authors: | dos Reis, C.V, Dutra, L.A, Gama, F.H, Mascarello, A, Azevedo, H, Guimaraes, C.R, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-24 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Vaccinia-related Kinase 1 (VRK1) bound to ACH471
To Be Published
|
|
2BU1
 
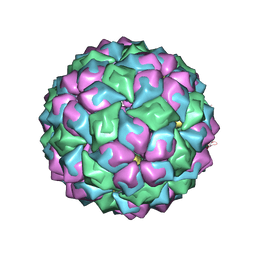 | | MS2-RNA HAIRPIN (5BRU -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *5BU*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
5VPP
 
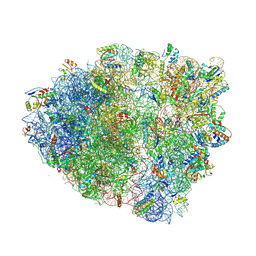 | | The 70S P-site tRNA SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8H6K
 
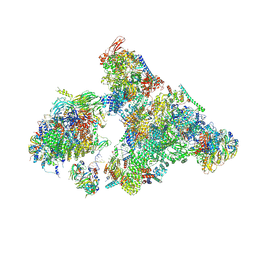 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
2BHI
 
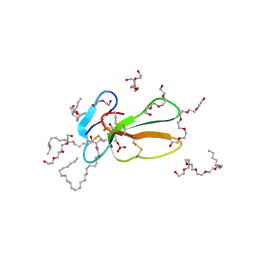 | | Crystal structure of Taiwan cobra cardiotoxin A3 complexed with sulfogalactoceramide | | Descriptor: | CYTOTOXIN 3, HEXAETHYLENE GLYCOL MONODECYL ETHER, SULFOGALACTOCERAMIDE | | Authors: | Wang, C.-H, Liu, J.-H, Wu, P.-L, Lee, S.-C, Hsiao, C.-D, Wu, W.-G. | | Deposit date: | 2005-01-12 | | Release date: | 2005-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycosphingolipid-Facilitated Membrane Insertion and Internalization of Cobra Cardiotoxin: The Sulfatide/Cardiotoxin Complex Structure in a Membrane-Like Environment Suggests a Lipid-Dependent Cell-Penetrating Mechanism for Membrane Binding Polypeptides.
J.Biol.Chem., 281, 2006
|
|
8H6L
 
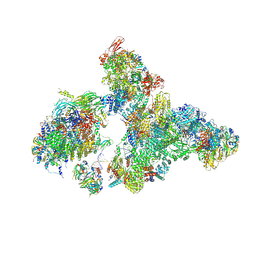 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
5VFC
 
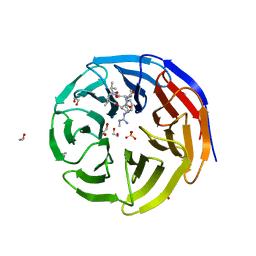 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
6X4C
 
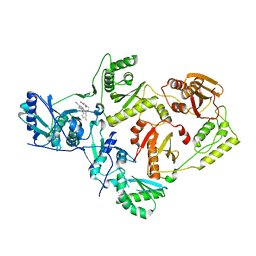 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-5,8-dimethyl-2-naphthonitrile (JLJ658), a Non-nucleoside Inhibitor | | Descriptor: | 7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5,8-dimethylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
2C3S
 
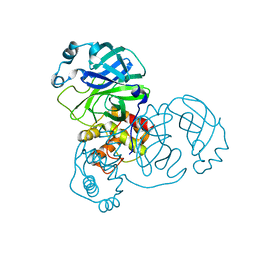 | | Structure Of Sars Cov Main Proteinase At 1.9 A (Ph6.5) | | Descriptor: | SARS COV 3C-LIKE PROTEINASE | | Authors: | Xu, T, Ooi, A, Lee, H.-C, Lescar, J. | | Deposit date: | 2005-10-12 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Sars Coronavirus Main Proteinase as an Active C2 Crystallographic Dimer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5VJX
 
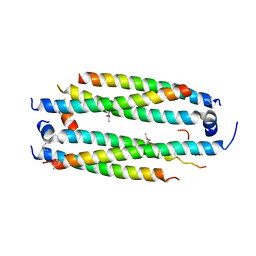 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
6X4A
 
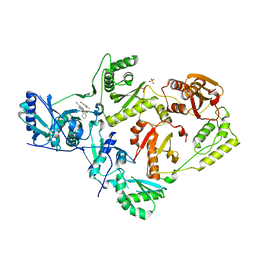 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
8I03
 
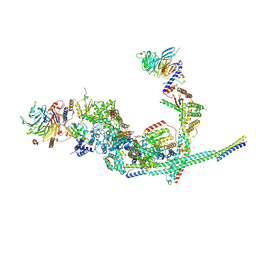 | | Cryo-EM structure of the SIN3L complex from S. pombe | | Descriptor: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
