6DBH
 
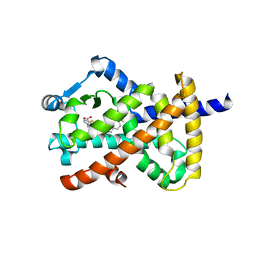 | | Crystal structure of PPAR gamma in complex with NMP422 | | Descriptor: | 4-{2-[(2,3-dioxo-1-pentyl-2,3-dihydro-1H-indol-5-yl)sulfanyl]ethyl}benzoic acid, CHLORIDE ION, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structure of PPAR gamma in complex with NMP422
To Be Published
|
|
4V9G
 
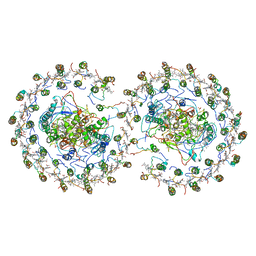 | | RC-LH1-PufX dimer complex from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Qian, P, Papiz, M.Z, Jackson, P.J, Brindley, A.A, Ng, I.W, Olsen, J.D, Dickman, M.J, Bullough, P.A, Hunter, C.N. | | Deposit date: | 2013-02-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (7.78 Å) | | Cite: | Three-Dimensional Structure of the Rhodobacter sphaeroides RC-LH1-PufX Complex: Dimerization and Quinone Channels Promoted by PufX.
Biochemistry, 52, 2013
|
|
4W8K
 
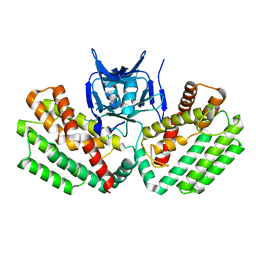 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
2BDV
 
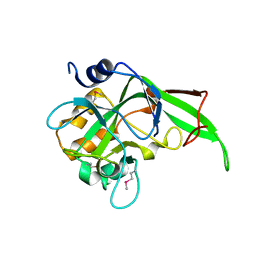 | | X-Ray Crystal Structure of Phage-related Protein BB2244 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium Target BoR24. | | Descriptor: | phage-related conserved hypothetical protein, BB2244 | | Authors: | Forouhar, F, Abashidze, M, Benach, J, Jayaraman, S, Janjua, H, Cooper, B, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the phage-related conserved hypothetical protein BB2244 from Bordetella bronchiseptica, Northeast Structural Genomics Target BoR24
To be Published
|
|
4V5I
 
 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LHE
 
 | | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne | | Descriptor: | CHLORIDE ION, GLYCEROL, GntR family Transcriptional regulator | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The crystal structure of the C-terminal domain of a GntR family transcriptional regulator from Bacillus anthracis str. Sterne
To be Published
|
|
3L5Z
 
 | | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Transcriptional regulator, ... | | Authors: | Chang, C, Hatzos, C, Feldmann, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of transcriptional regulator, GntR family from Bacillus cereus
To be Published
|
|
3LMP
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-N-(1-naphthylmethyl)-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide of Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel selective PPARgamma modulator from (-)-Cercosporamide derivatives
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7BOG
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
1JFT
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PHOSPHATE ION, ... | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
2VUK
 
 | |
7T8O
 
 | | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes | | Descriptor: | Lmo0753 protein, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Crp/Fnr Family Transcriptional Regulator from Listeria monocytogenes
To Be Published
|
|
4V5D
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
5NWY
 
 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
5O2R
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide Api137 bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7BL1
 
 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
8T9D
 
 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8TRH
 
 | | The IDRc bound human core Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TQW
 
 | | Structure of human transcriptional Mediator complex | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-08 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
5NCO
 
 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2018-03-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
1DQ3
 
 | |
5NIO
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-butyl-4-methyl-piperidine-1-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NIZ
 
 | | EthR complex | | Descriptor: | 1-[3-[5-[(1~{R},2~{R})-2-methylcyclopropyl]furan-2-yl]propanoyl]piperidine-4-carboxamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
3CF2
 
 | | Structure of P97/vcp in complex with ADP/AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
