1POS
 
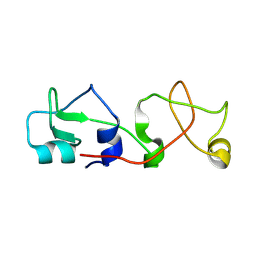 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1PRG
 
 | |
1PU2
 
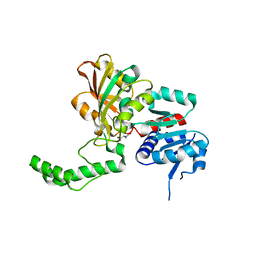 | | Crystal Structure of the K246R Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-06-23 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PTT
 
 | |
1PUB
 
 | |
1Q1S
 
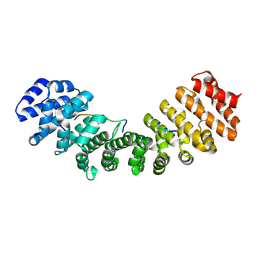 | | Mouse Importin alpha- phosphorylated SV40 CN peptide complex | | Descriptor: | Importin alpha-2 subunit, Large T antigen | | Authors: | Fontes, M.R.M, Teh, T, Toth, G, John, A, Pavo, I, Jans, D.A, Kobe, B. | | Deposit date: | 2003-07-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha
Biochem.J., 375, 2003
|
|
1Q2Y
 
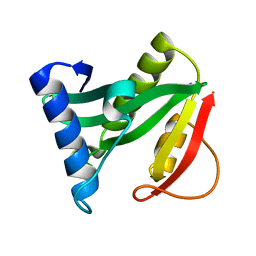 | | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily fold | | Descriptor: | similar to hypothetical proteins | | Authors: | Fedorov, A.A, Ramagopal, U.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-27 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily
To be Published
|
|
1PX8
 
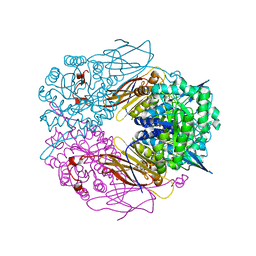 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1PKM
 
 | |
1PYX
 
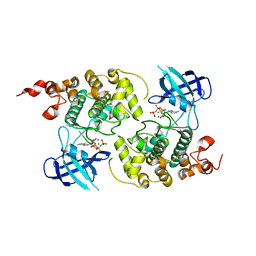 | | GSK-3 Beta complexed with AMP-PNP | | Descriptor: | Glycogen synthase kinase-3 beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors
J.Mol.Biol., 333, 2003
|
|
1PL8
 
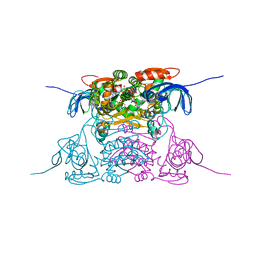 | | human SDH/NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION, human sorbitol dehydrogenase | | Authors: | Pauly, T.A, Ekstrom, J.L, Beebe, D.A, Chrunyk, B, Cunningham, D, Griffor, M, Kamath, A, Lee, S.E, Madura, R, Mcguire, D, Subashi, T, Wasilko, D, Watts, P, Mylari, B.L, Oates, P.J, Adams, P.D, Rath, V.L. | | Deposit date: | 2003-06-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic and kinetic studies of human sorbitol dehydrogenase.
Structure, 11, 2003
|
|
1PNE
 
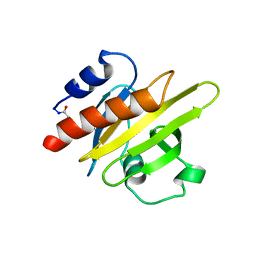 | | CRYSTALLIZATION AND STRUCTURE DETERMINATION OF BOVINE PROFILIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | PROFILIN | | Authors: | Cedergren-Zeppezauer, E.S, Goonesekere, N.C.W, Rozycki, M.D, Myslik, J.C, Dauter, Z, Lindberg, U, Schutt, C.E. | | Deposit date: | 1995-05-05 | | Release date: | 1995-07-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and structure determination of bovine profilin at 2.0 A resolution.
J.Mol.Biol., 240, 1994
|
|
1PNK
 
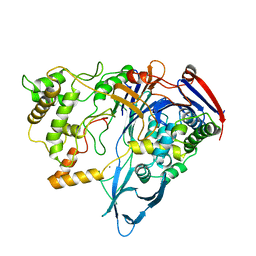 | |
1PPI
 
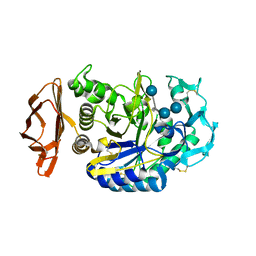 | | THE ACTIVE CENTER OF A MAMMALIAN ALPHA-AMYLASE. THE STRUCTURE OF THE COMPLEX OF A PANCREATIC ALPHA-AMYLASE WITH A CARBOHYDRATE INHIBITOR REFINED TO 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Qian, M, Haser, R, Payan, F. | | Deposit date: | 1994-02-22 | | Release date: | 1995-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active center of a mammalian alpha-amylase. Structure of the complex of a pancreatic alpha-amylase with a carbohydrate inhibitor refined to 2.2-A resolution.
Biochemistry, 33, 1994
|
|
1Q1Y
 
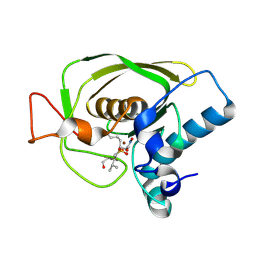 | | Crystal Structures of Peptide Deformylase from Staphylococcus aureus Complexed with Actinonin | | Descriptor: | ACTINONIN, Peptide deformylase, ZINC ION | | Authors: | Yoon, H.J, Lee, S.K, Kim, H.L, Kim, H.W, Kim, H.W, Lee, J.Y, Mikami, B, Suh, S.W. | | Deposit date: | 2003-07-23 | | Release date: | 2004-07-23 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
1O5T
 
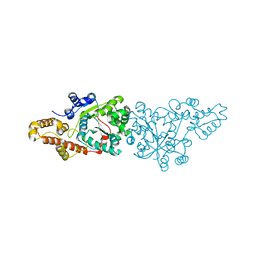 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
1O9Q
 
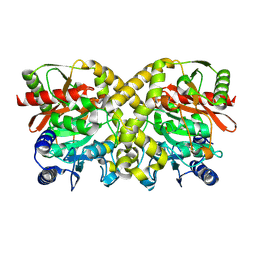 | |
1OAS
 
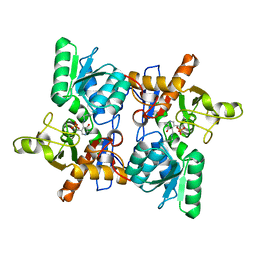 | | O-ACETYLSERINE SULFHYDRYLASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Rao, G.S.J, Hohenester, E, Schnackerz, K.D, Cook, P.F, Jansonius, J.N. | | Deposit date: | 1999-01-29 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium.
J.Mol.Biol., 283, 1998
|
|
1OCK
 
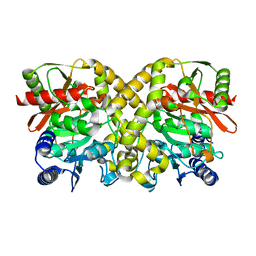 | |
1ODI
 
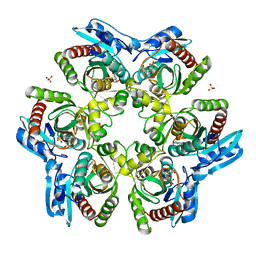 | |
1NMB
 
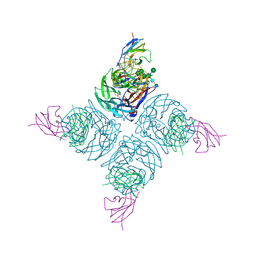 | | THE STRUCTURE OF A COMPLEX BETWEEN THE NC10 ANTIBODY AND INFLUENZA VIRUS NEURAMINIDASE AND COMPARISON WITH THE OVERLAPPING BINDING SITE OF THE NC41 ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB NC10, ... | | Authors: | Malby, R.L, Tulip, W.R, Colman, P.M. | | Deposit date: | 1995-01-17 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Structure, 2, 1994
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
1NRX
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ and NAD | | Descriptor: | 3-dehydroquinate synthase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-26 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NSC
 
 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1NUA
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
