7BW1
 
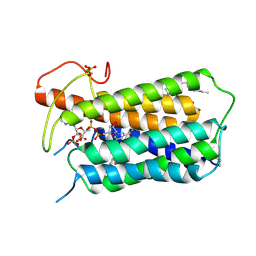 | | Crystal structure of Steroid 5-alpha-reductase 2 in complex with Finasteride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase 2, SULFATE ION, ... | | Authors: | Xiao, Q, Zhang, C, Wei, Z. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human steroid 5 alpha-reductase 2 with anti-androgen drug finasteride.
Res Sq, 2020
|
|
6ZXN
 
 | |
7N5Z
 
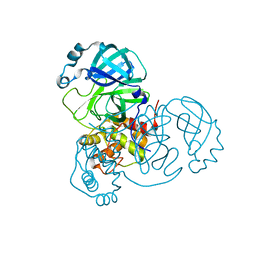 | | SARS-CoV-2 Main protease C145S mutant | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7V61
 
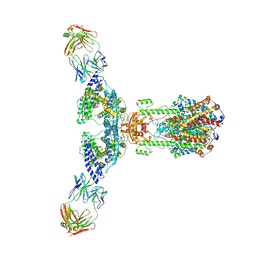 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-08-18 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
7MZK
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZJ
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 93 | | Descriptor: | GLYCEROL, PDI 93 heavy chain, PDI 93 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | Descriptor: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZF
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 37 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZM
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 215 | | Descriptor: | ISOPROPYL ALCOHOL, PDI 215 heavy chain, PDI 215 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZL
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 210 heavy chain, PDI 210 light chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | Descriptor: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZN
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 231 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 231 heavy chain, PDI 231 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7NL7
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7N5H
 
 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7TV0
 
 | |
7TUQ
 
 | |
7WHC
 
 | | Crystal structure of SARS-CoV-2 3CLpro catalytic domain | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Shin, D.H, Jo, S.R. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro.
Int J Mol Sci, 23, 2022
|
|
7U0P
 
 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7O7F
 
 | | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist | | Descriptor: | C-C chemokine receptor type 5, C-C motif chemokine 5, Fab antibody fragment heavy chain, ... | | Authors: | Isaikina, P, Tsai, C.-J, Dietz, N.B, Pamula, F, Goldie, K.N, Schertler, G.F.X, Maier, T, Stahlberg, H, Deupi, X, Grzesiek, S. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist.
Sci Adv, 7, 2021
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7ON5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7OZV
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZU
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
