7VSO
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with HAD16 Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2-[[(2R)-2-(3-bromanyl-5-iodanyl-phenyl)carbonyloxy-3-tetradecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, ... | | Authors: | Mizohata, E, Nakane, T, Hanashima, S. | | Deposit date: | 2021-10-27 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins.
Membranes (Basel), 11, 2021
|
|
6ME9
 
 | | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon | | Descriptor: | N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6MF0
 
 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
8HL6
 
 | |
8EFH
 
 | | Helical reconstruction of the human cardiac actin-tropomyosin-myosin complex in complex with ADP-Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Lehman, W, Rynkiewicz, M.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes linked to ADP release from human cardiac myosin bound to actin-tropomyosin.
J.Gen.Physiol., 155, 2023
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7VEL
 
 | | Crystal structure of Phytolacca americana UGT3 with UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8HL7
 
 | |
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7V33
 
 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VEJ
 
 | | Crystal structure of Phytolacca americana UGT3 with kaempferol and UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VEK
 
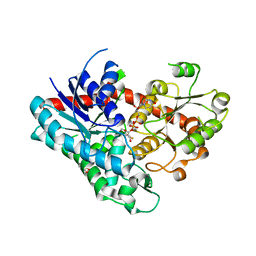 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7V32
 
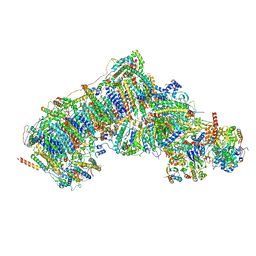 | | Deactive state complex I from rotenone dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6LTN
 
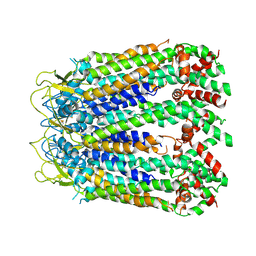 | | cryo-EM structure of C-terminal truncated human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
7V3M
 
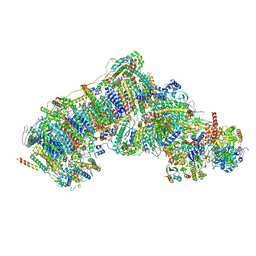 | | Deactive state complex I from rotenone-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8EPA
 
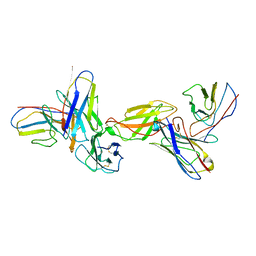 | |
7V31
 
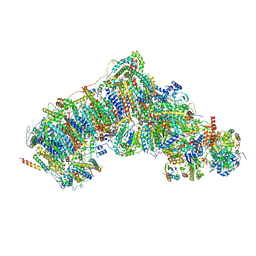 | | Active state complex I from rotenone dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8F6H
 
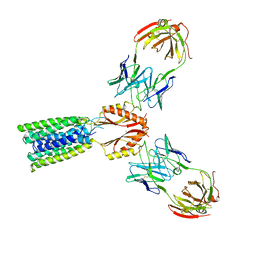 | | Cryo-EM structure of a Zinc-loaded asymmetrical TMD D70A mutant of the YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
6LZ0
 
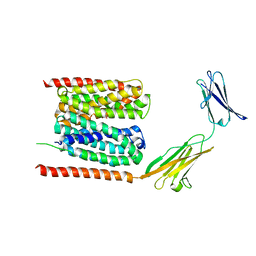 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
8GY2
 
 | | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | | Descriptor: | Alcohol dehydrogenase (quinone), cytochrome c subunit, dehydrogenase subunit, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
6M23
 
 | | Overall structure of KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
