6KGN
 
 | | LSD1-CoREST-S2116 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
4DM6
 
 | | Crystal structure of RARb LBD homodimer in complex with TTNPB | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Nuclear receptor coactivator 1, Retinoic acid receptor beta | | Authors: | Osz, J, Brelivet, Y, Peluso-Iltis, C, Cura, V, Eiler, S, Ruff, M, Bourguet, W, Rochel, N, Moras, D. | | Deposit date: | 2012-02-07 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6SWA
 
 | | Mus musculus brain neocortex ribosome 60S bound to Ebp1 | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kraushar, M.L, Sprink, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-30 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Protein Synthesis in the Developing Neocortex at Near-Atomic Resolution Reveals Ebp1-Mediated Neuronal Proteostasis at the 60S Tunnel Exit.
Mol.Cell, 81, 2021
|
|
8JF5
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAS1440 | | Descriptor: | 4-[5-[(3~{R})-3-azanylpyrrolidin-1-yl]carbonyl-2-[2-fluoranyl-4-(2-methyl-2-oxidanyl-propyl)phenyl]phenyl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Fukushima, H, Machida, T, Yamashita, S, Suzuki, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | TAS1440, a histone H3 competitive LSD1 inhibitor, activates both TGF-beta and notch signaling pathways via INSM1 dissociation in neuroendocrine small cell lung cancer
To Be Published
|
|
8OEW
 
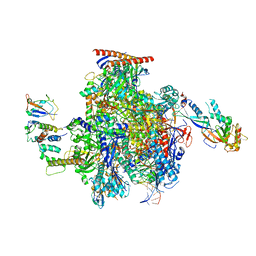 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEV
 
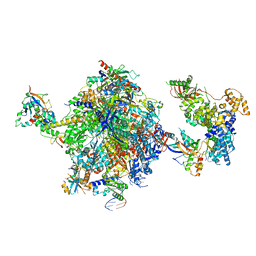 | | Structure of the mammalian Pol II-SPT6-Elongin complex, lacking ELOA latch (composite structure, structure 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6S35
 
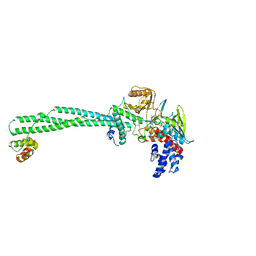 | | LSD1/CoREST1 complex with macrocyclic peptide inhibitor | | Descriptor: | ALA-ARG-(D)LYS-MET-GLN-GLU-ALA-ARG-LYS-SER-THR, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Talibov, V.O, Dobritzsch, D. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Macrocyclic Peptides Uncover a Novel Binding Mode for Reversible Inhibitors of LSD1.
Acs Omega, 5, 2020
|
|
6W9K
 
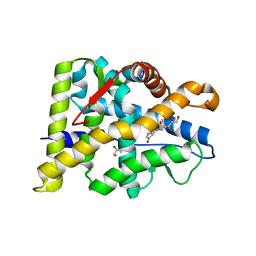 | |
6W9M
 
 | |
6W9L
 
 | | Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with deacetylated deflazacort and PGC1a coregulator fragment | | Descriptor: | (4aR,4bS,5S,6aS,6bS,9aR,10aS,10bS)-5-hydroxy-6b-(hydroxyacetyl)-4a,6a,8-trimethyl-4a,4b,5,6,6a,6b,9a,10,10a,10b,11,12-dodecahydro-2H-naphtho[2',1':4,5]indeno[1,2-d][1,3]oxazol-2-one, GLYCEROL, Glucocorticoid Receptor, ... | | Authors: | Liu, X, Ortlund, E.A. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Disruption of a key ligand-H-bond network drives dissociative properties in vamorolone for Duchenne muscular dystrophy treatment.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5H6Q
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
4CZZ
 
 | | Histone demethylase LSD1(KDM1A)-CoREST3 Complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 3 | | Authors: | Barrios, A.P, Gomez, A.V, Saez, J.E, Ciossani, G, Toffolo, E, Battaglioli, E, Mattevi, A, Andres, M.E. | | Deposit date: | 2014-04-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Properties of Transcriptional Complexes Formed by the Corest Family.
Mol.Cell.Biol., 34, 2014
|
|
3TX7
 
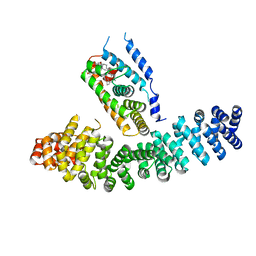 | | Crystal structure of LRH-1/beta-catenin complex | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Catenin beta-1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Yumoto, F, Fletterick, R. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis of coactivation of liver receptor homolog-1 by beta-catenin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8HY0
 
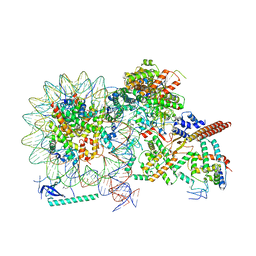 | |
8HXY
 
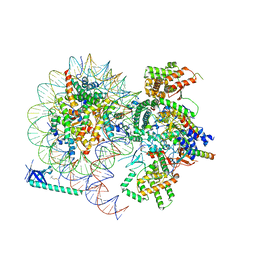 | |
6AN1
 
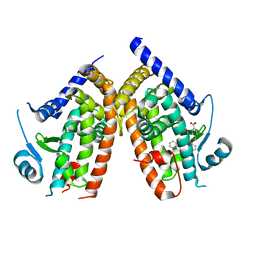 | | Crystal structure of the complex between PPARgamma LBD and the ligand AM-879 | | Descriptor: | 4-({2-[(1,3-dioxo-1,3-dihydro-2H-inden-2-ylidene)methyl]phenoxy}methyl)benzoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Veras, H, Figueira, A.C, le Maire, A. | | Deposit date: | 2017-08-11 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Screening for PPAR Non-Agonist Ligands Followed by Characterization of a Hit, AM-879, with Additional No-Adipogenic and cdk5-Mediated Phosphorylation Inhibition Properties.
Front Endocrinol (Lausanne), 9, 2018
|
|
8JAR
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAQ
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAU
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1MM2
 
 | | Solution structure of the 2nd PHD domain from Mi2b | | Descriptor: | Mi2-beta, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
6C9L
 
 | | MEF2B Apo Protein Structure | | Descriptor: | Myocyte-specific enhancer factor 2B | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2018-01-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Apo MEF2B Reveals New Insights in DNA Binding and Cofactor Interaction.
Biochemistry, 57, 2018
|
|
5VTB
 
 | | Crystal structure of RBBP4 bound to BCL11a peptide | | Descriptor: | B-cell lymphoma/leukemia 11A, GLYCEROL, Histone-binding protein RBBP4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes.
J. Biol. Chem., 293, 2018
|
|
