6XK8
 
 | |
6EA5
 
 | | Structure of BDBV GPcl in complex with the pan-ebolavirus mAb ADI-15878 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-15878 Fab Heavy Chain, ... | | Authors: | King, L.B, West, B.R, Moyer, C.L, Fusco, M.L, Milligan, J.C, Hui, S, Saphire, E.O. | | Deposit date: | 2018-08-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structural Basis of Pan-Ebolavirus Neutralization by a Human Antibody against a Conserved, yet Cryptic Epitope.
MBio, 9, 2018
|
|
7JPJ
 
 | | Crystal Structure of the essential dimeric LYSA from Phaeodactylum tricornutum | | Descriptor: | D-LYSINE, Diaminopimelate decarboxylase, SULFATE ION | | Authors: | Fedorov, E, Belinski, V.A, Brunson, J.K, Almo, S.C, Dupont, C.L, Ghosh, A. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Phaeodactylum tricornutum diaminopimelate decarboxylase was acquired via horizontal gene transfer from bacteria and displays substrate promiscuity
Biorxiv, 2020
|
|
5U1E
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Putative gentamicin methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5F5J
 
 | |
6XK4
 
 | |
6IO6
 
 | |
6EAE
 
 | |
7ZVP
 
 | |
6B8B
 
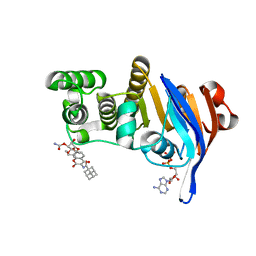 | | E. coli LptB in complex with ADP and a novobiocin derivative | | Descriptor: | (3s,5s,7s)-N-{7-[(3-O-carbamoyl-6-deoxy-5-methyl-4-O-methyl-beta-D-gulopyranosyl)oxy]-4-hydroxy-8-methyl-2-oxo-2H-1-ben zopyran-3-yl}tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Mandler, M.D, Owens, T.W, Lazarus, M.B, May, J.M, Kahne, D.K. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
8QXX
 
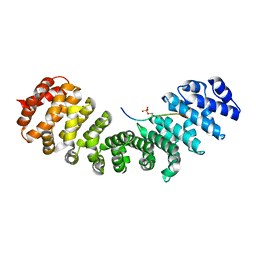 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
6EAJ
 
 | |
6BBT
 
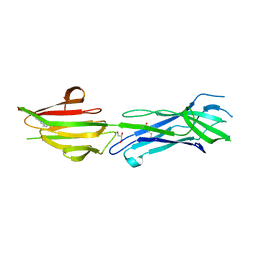 | | Structure of the major pilin protein (T-13) from Streptococcus pyogenes serotype GAS131465 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Young, P.G, Baker, E.N, Moreland, N.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
8QPC
 
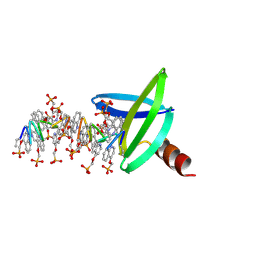 | |
7ZUN
 
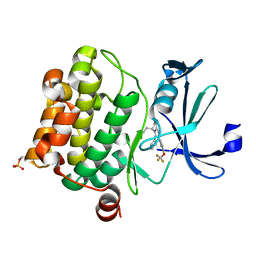 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone compound | | Descriptor: | (4~{S})-4-(2-azanylethyl)-6-phenyl-7-[3-(trifluoromethyloxy)phenyl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1-ol, Isoform 2 of Serine/threonine-protein kinase pim-1 | | Authors: | Casale, E. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Chirality, 34, 2022
|
|
8QLT
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR30 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[3-(2-oxidanylidenepyrrolidin-1-yl)propylamino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6XJN
 
 | |
8A3G
 
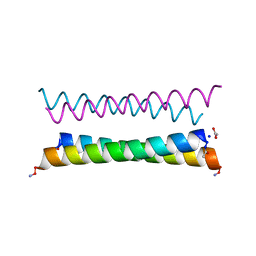 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 4 heptad repeats, apCC-Tet* | | Descriptor: | ACETATE ION, SODIUM ION, apCC-Tet* | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
6XL4
 
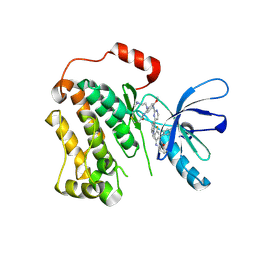 | | EGFR(T790M/V948R) in complex with AZD9291 and DDC4002 | | Descriptor: | 10-benzyl-8-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors.
Nat Commun, 13, 2022
|
|
6EBB
 
 | |
5F5W
 
 | |
8QXW
 
 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
5EWA
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor L-VC26 | | Descriptor: | (3~{R},5~{R},7~{a}~{S})-2,2-dimethyl-5-(sulfanylmethyl)-3,5,7,7~{a}-tetrahydro-[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Kosmopoulou, M, Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6AZ5
 
 | |
6XK7
 
 | |
