1DKJ
 
 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1E7L
 
 | | Endonuclease VII (EndoVII) N62D mutant from phage T4 | | Descriptor: | RECOMBINATION ENDONUCLEASE VII, SULFATE ION, ZINC ION | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
1E6D
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH TRP M115 REPLACED WITH PHE (CHAIN M, WM115F) PHE M197 REPLACED WITH ARG (CHAIN M, FM197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ridge, J.P, Fyfe, P.K, McAuley, K.E, Van Brederode, M.E, Robert, B, Van Grondelle, R, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2000-08-11 | | Release date: | 2000-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Examination of How Structural Changes Can Affect the Rate of Electron Transfer in a Mutated Bacterial Photoreaction Centre
Biochem.J., 351, 2000
|
|
2NCK
 
 | |
8V9J
 
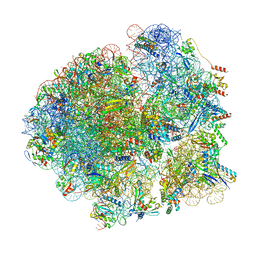 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S14, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8UKT
 
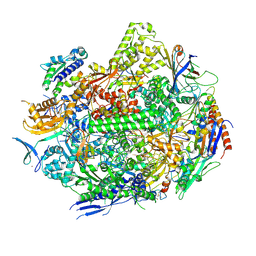 | | RNA polymerase II elongation complex with Fapy-dG lesion with AMP added | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
8TNC
 
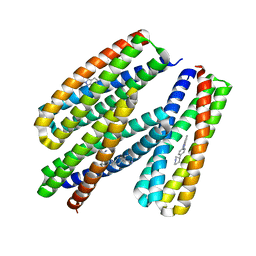 | |
8VGC
 
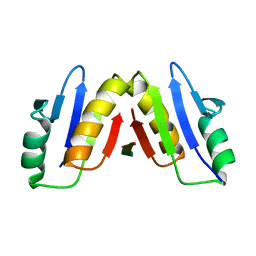 | | Complex of ExbD with D-box peptide: Orthorhombic form | | Descriptor: | Biopolymer transport protein ExbD, GLN-PRO-ILE-SER-VAL-THR-MET-VAL-THR-PRO | | Authors: | Loll, P.J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and structural characterization of the D-box, a conserved TonB motif that couples an inner-membrane motor to outer-membrane transport.
J.Biol.Chem., 300, 2024
|
|
8WZJ
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9BH6
 
 | |
9B35
 
 | | Ligand-binding and transmembrane domains of kainate receptor GluK2 in the open state, a complex with agonist glutamate and positive allosteric modulator BPAM344 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor ionotropic, kainate 2 | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
8VGD
 
 | | Complex of ExbD with D-box peptide: Tetragonal form | | Descriptor: | Biopolymer transport protein ExbD, GLN-PRO-ILE-SER-VAL-THR-MET-VAL-THR | | Authors: | Loll, P.J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and structural characterization of the D-box, a conserved TonB motif that couples an inner-membrane motor to outer-membrane transport.
J.Biol.Chem., 300, 2024
|
|
8W4Q
 
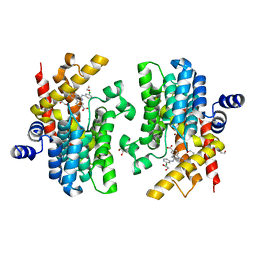 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W9Y
 
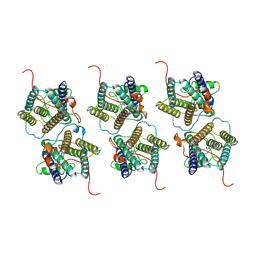 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | Descriptor: | Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9ILB
 
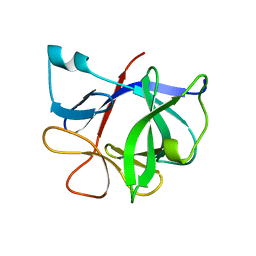 | | HUMAN INTERLEUKIN-1 BETA | | Descriptor: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | Authors: | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8YZ7
 
 | |
8XLM
 
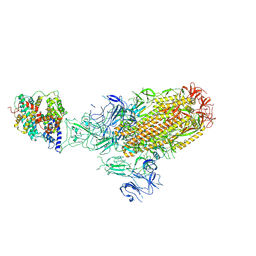 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8W4R
 
 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8UUE
 
 | |
8WUY
 
 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
8YZK
 
 | | Orphan receptor GPRC5D in complex with scFv150-18 | | Descriptor: | Soluble cytochrome b562,G-protein coupled receptor family C group 5 member D, scFv | | Authors: | Yan, P, Lin, X, Xu, F. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The binding mechanism of an anti-multiple myeloma antibody to the human GPRC5D homodimer.
Nat Commun, 15, 2024
|
|
8Y8Z
 
 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8W78
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
