9G6K
 
 | | LSU structure derived from the LSU sample of the mitoribosome from T. gondii. | | Descriptor: | AMP-dependent synthetase/ligase domain-containing protein, AP2 domain transcription factor AP2IV-1, AP2 domain transcription factor AP2VIIb-2, ... | | Authors: | Rocha, R.E.O, Barua, S, Boissier, F, Nguyen, T.T, Hashem, Y. | | Deposit date: | 2024-07-18 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Apicomplexan mitoribosome from highly fragmented rRNAs to a functional machine.
Nat Commun, 15, 2024
|
|
6THD
 
 | | Multiple Genomic RNA-Coat Protein Contacts Play Vital Roles in the Assembly of Infectious Enterovirus-E | | Descriptor: | Genome polyprotein, MYRISTIC ACID, SULFATE ION | | Authors: | Chandler-Bostock, R, Mata, C.P, Bingham, R, Dykeman, E.J, Meng, B, Tuthill, T.J, Rowlands, D.J, Ranson, N.A, Twarock, R, Stockley, P.G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts.
Plos Pathog., 16, 2020
|
|
6THN
 
 | | Multiple Genomic RNA-Coat Protein Contacts Play Vital Roles in the Assembly of Infectious Enterovirus-E symmetry expansion+2fold focused classification | | Descriptor: | Genome polyprotein, MYRISTIC ACID, RNA Peak 9 Bernoulli Plot, ... | | Authors: | Chandler-Bostock, R, Mata, C.P, Bingham, R, Dykeman, E.J, Meng, B, Tuthill, T.J, Rowlands, D.J, Ranson, N.A, Twarock, R, Stockley, P.G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts.
Plos Pathog., 16, 2020
|
|
6NY6
 
 | | Structure of dimeric Escherichia coli toxin YoeB bound to the Thermus thermophilus 30S ribosome | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Pavelich, I.J, Hoffer, E.D, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability.
Nucleic Acids Res., 47, 2019
|
|
1LLR
 
 | | CHOLERA TOXIN B-PENTAMER WITH LIGAND BMSC-0012 | | Descriptor: | 3-AMINO-4-{3-[2-(2-PROPOXY-ETHOXY)-ETHOXY]-PROPYLAMINO}-CYCLOBUT-3-ENE-1,2-DIONE, 5-aminocarbonyl-3-nitrophenyl alpha-D-galactopyranoside, CHOLERA TOXIN B SUBUNIT | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Characterization and crystal structure of a high-affinity pentavalent receptor-binding inhibitor for cholera toxin and E. coli heat-labile enterotoxin.
J.Am.Chem.Soc., 124, 2002
|
|
2UZB
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]-2-FURYL}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7RB7
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
2UZL
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]FURAN-2-YL}-2-(TRIFLUOROMETHYL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2UZD
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]FURAN-2-YL}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6M5Q
 
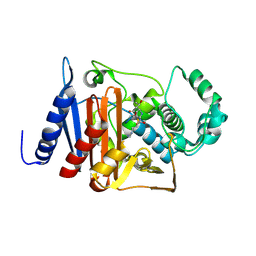 | | A class C beta-lactamase mutant - Y150F | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
9ECT
 
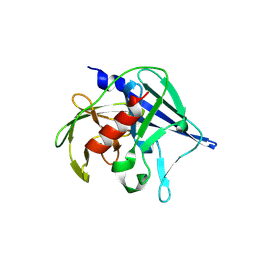 | |
6FDQ
 
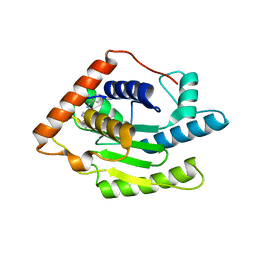 | |
6CJW
 
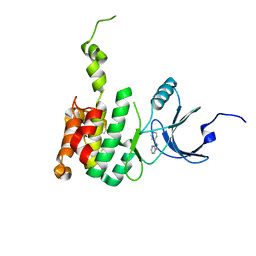 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-4-yl)imidazo[1,2-b]pyridazine, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6V0X
 
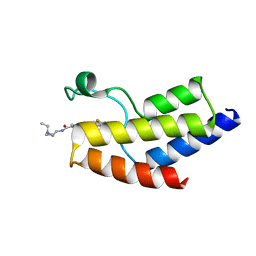 | | Crystal structure of the bromodomain of human BRD9 bound to sunitinib | | Descriptor: | Bromodomain-containing protein 9, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Karim, M.R, Chan, A, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
7KJX
 
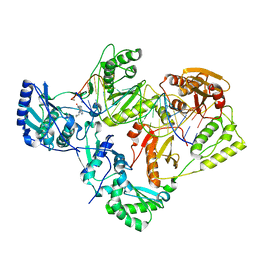 | | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
3OEO
 
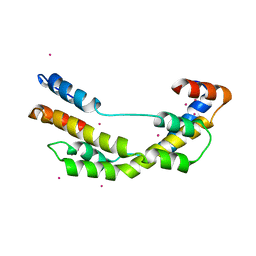 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
1NI7
 
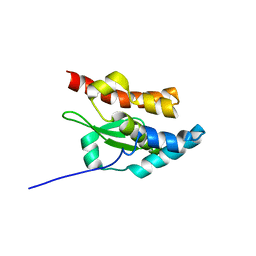 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
8UXQ
 
 | |
5VP2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with madumycin II and bound to mRNA and A-, P- and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Osterman, I.A, Khabibullina, N.F, Komarova, E.S, Kasatsky, P, Kartsev, V.G, Bogdanov, A.A, Dontsova, O.A, Konevega, A.L, Sergiev, P.V, Polikanov, Y.S. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-28 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state.
Nucleic Acids Res., 45, 2017
|
|
5W4K
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
6H4N
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
5NAL
 
 | | The crystal structure of inhibitor-15 covalently bound to PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}1-(cyclohexylmethyl)-~{N}4-cyclopentyl-~{N}1-[(~{Z})-4-[(~{E})-methyliminomethyl]-5-oxidanyl-hex-4-enyl]benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent Protein Labeling at Glutamic Acids.
Cell Chem Biol, 24, 2017
|
|
8A0L
 
 | | Tubulin-CW1-complex | | Descriptor: | (3~{S},4~{R},8~{S},10~{S},12~{S},14~{S})-14-[(~{Z},4~{R})-4-(hydroxymethyl)hex-2-en-2-yl]-4,12-dimethoxy-9,9-dimethyl-3,8,10-tris(oxidanyl)-1-oxacyclotetradecan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Diaz, J.F, Steinmetz, M.O, Oliva, M.A. | | Deposit date: | 2022-05-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9981 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
6HEF
 
 | | Room temperature structure of the (SR)Ca2+-ATPase Ca2-E1-CaAMPPCP form | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Hjorth-Jensen, S, Sorensen, T.L.M, Oksanen, E, Andersen, J.L, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.538 Å) | | Cite: | Membrane-protein crystals for neutron diffraction.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2UZE
 
 | | Crystal structure of human CDK2 complexed with a thiazolidinone inhibitor | | Descriptor: | 4-{5-[(Z)-(2-IMINO-4-OXO-1,3-THIAZOLIDIN-5-YLIDENE)METHYL]FURAN-2-YL}BENZOIC ACID, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent Cdk2 Inhibitor with a Novel Binding Mode, Using Virtual Screening and Initial, Structure-Guided Lead Scoping.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
