2J0D
 
 | |
2R97
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2RHB
 
 | | Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Palaninathan, S, Bhardwaj, K, Alcantara, J.M.O, Guarino, L, Yi, L.L, Kao, C.C, Sacchettini, J. | | Deposit date: | 2007-10-08 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.
J.Biol.Chem., 283, 2008
|
|
2RG1
 
 | | Crystal structure of E. coli WrbA apoprotein | | Descriptor: | CHLORIDE ION, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Lapkouski, M, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2R8G
 
 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DT)-3', 5'-D(*DTP*DCP*DAP*DCP*(P)P*DGP*DAP*DAP*DAP*DTP*DCP*DCP*DTP*DTP*DCP*DCP*DCP*DCP*DC)-3', ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
2I57
 
 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
3IBD
 
 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
3IH7
 
 | |
3II4
 
 | |
2NNI
 
 | | CYP2C8dH complexed with montelukast | | Descriptor: | Cytochrome P450 2C8, MONTELUKAST, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
6EB0
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, ACETATE ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Yan, Y, Rose, J. | | Deposit date: | 2018-08-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
2NNH
 
 | | CYP2C8dH complexed with 2 molecules of 9-cis retinoic acid | | Descriptor: | (9cis)-retinoic acid, Cytochrome P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
2IJ7
 
 | | Structure of Mycobacterium tuberculosis CYP121 in complex with the antifungal drug fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roujeinikova, A, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode.
J.Biol.Chem., 281, 2006
|
|
2J4W
 
 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
2J5L
 
 | | Structure of a Plasmodium falciparum apical membrane antigen 1-Fab F8. 12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
2NOL
 
 | | Structure of catalytically inactive human 8-oxoguanine glycosylase distal crosslink to oxoG DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Radom, C.T, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NNJ
 
 | | CYP2C8dH complexed with felodipine | | Descriptor: | Cytochrome P450 2C8, FELODIPINE, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
6FAH
 
 | | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction | | Descriptor: | Caffeyl-CoA reductase-Etf complex subunit CarC, Caffeyl-CoA reductase-Etf complex subunit CarD, Caffeyl-CoA reductase-Etf complex subunit CarE, ... | | Authors: | Demmer, J.K, Bertsch, J, Oeppinger, C, Wohlers, H, Kayastha, K, Demmer, U, Ermler, U, Mueller, V. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction.
FEBS Lett., 592, 2018
|
|
2NOH
 
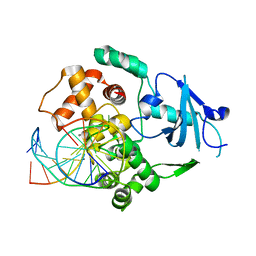 | | Structure of catalytically inactive Q315A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOZ
 
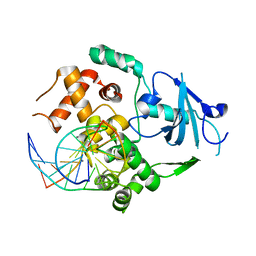 | | Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOF
 
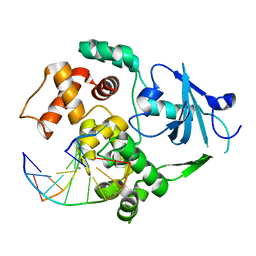 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
6J31
 
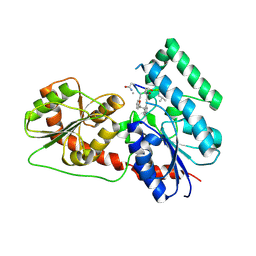 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
2NOI
 
 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOB
 
 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
6J32
 
 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | Kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
