5JQS
 
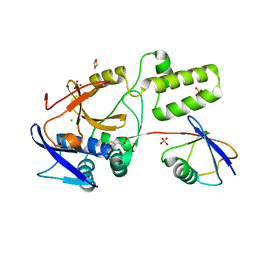 | |
6DH2
 
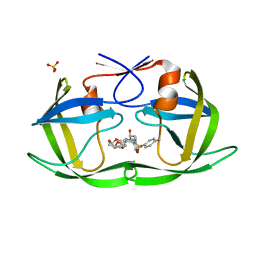 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
5EI2
 
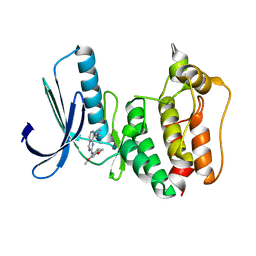 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
6DH4
 
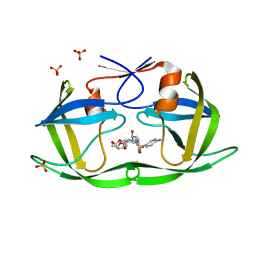 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
8DAF
 
 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
8AYF
 
 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
5JCU
 
 | |
5ES2
 
 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
6DH0
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH8
 
 | | Crystal structure of HIV-1 Protease NL4-3 I50V Mutant in complex with UMass6 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
6DH1
 
 | | Crystal structure of HIV-1 Protease NL4-3 I84V Mutant in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
5IWG
 
 | | HDAC2 WITH LIGAND BRD4884 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
6X5S
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3'-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A3'-Asn, Alpha-(1,6)-fucosyltransferase, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
5EI6
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-5-(1-methylpyrazol-4-yl)isoquinolin-3-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
7AP2
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound LeuS7HMDDA | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
5WCI
 
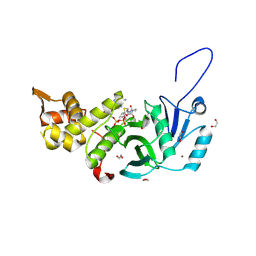 | | Human MYST histone acetyltransferase 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Zheng, Y.G, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Human MYST histone acetyltransferase 1
to be published
|
|
6HX1
 
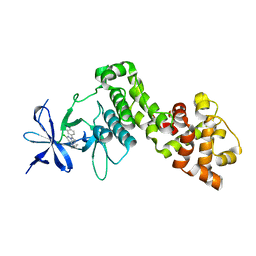 | | IRE1 ALPHA IN COMPLEX WITH imidazo[1,2-b]pyridazin-8-amine compound 2 | | Descriptor: | 6-chloranyl-~{N}-(cyclopropylmethyl)-3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Augustin, M.A, Krapp, S, Bayliss, R, Collins, I. | | Deposit date: | 2018-10-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
7AYH
 
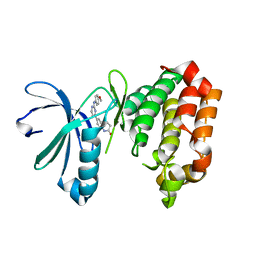 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
5NRD
 
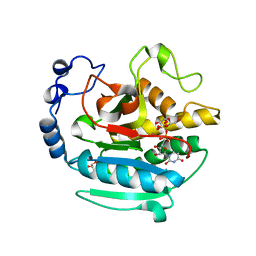 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7C7D
 
 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7LVI
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 ISOFORM 1 COMPLEXED WITH LIGAND (2R)-2-AMINO-N-[3-METHOXY-4- (1,3-OXAZOL-5-YL)PHENYL]-4-METHYLPENTANAMIDE | | Descriptor: | (2R,3R)-N-[3-methoxy-4-(1,3-oxazol-5-yl)phenyl]-3-(propan-2-yl)piperidine-2-carboxamide, 5-[(4-aminopiperidin-1-yl)methyl]-N-{3-[5-(propan-2-yl)-1,3,4-thiadiazol-2-yl]phenyl}pyrrolo[2,1-f][1,2,4]triazin-4-amine, AP2-associated protein kinase 1, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery, Structure-Activity Relationships, and In Vivo Evaluation of Novel Aryl Amides as Brain Penetrant Adaptor Protein 2-Associated Kinase 1 (AAK1) Inhibitors for the Treatment of Neuropathic Pain.
J.Med.Chem., 64, 2021
|
|
7SNP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-(2-morpholinoethoxy)phenoxy)phenyl)acrylonitrile (JLJ530) | | Descriptor: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-[2-(morpholin-4-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
5MX7
 
 | | 1a,20S-dihydroxyvitamin D3 VDR complex | | Descriptor: | 1a,20S-dihydroxyvitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2017-01-21 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Sci Rep, 7, 2017
|
|
7AP1
 
 | | Klebsiella pneumoniae Seryl-tRNA synthetase in Complex with Compound SerS7HMDDA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine-tRNA ligase, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
6DGY
 
 | | Crystal structure of HIV-1 Protease NL4-3 WT in complex with UMass1 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
