2ALP
 
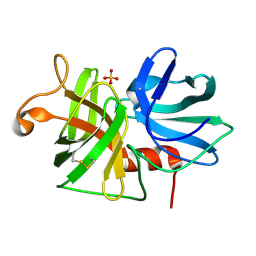 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
6CGV
 
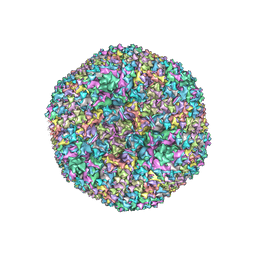 | | Revised crystal structure of human adenovirus | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Natchiar, S.K, Venkataraman, S, Nemerow, G.R, Reddy, V.S. | | Deposit date: | 2018-02-21 | | Release date: | 2018-04-25 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Revised Crystal Structure of Human Adenovirus Reveals the Limits on Protein IX Quasi-Equivalence and on Analyzing Large Macromolecular Complexes.
J. Mol. Biol., 430, 2018
|
|
2BE5
 
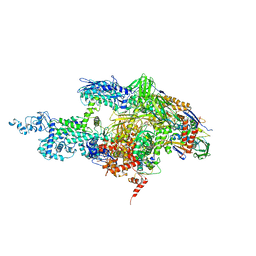 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
2PRQ
 
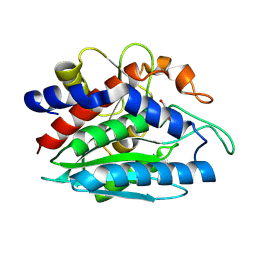 | | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, COBALT (II) ION | | Authors: | Munih, P, Moulin, A, Stamper, C.C, Bennet, B, Ringe, D, Petsko, G.A, Holz, R.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica.
J.Inorg.Biochem., 101, 2007
|
|
5CX7
 
 | | Crystal Structure of PduOC:Heme Complex | | Descriptor: | ATP:cob(I)alamin adenosyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Geremia, S, Hickey, N, Ortiz de Orue Lucana, D. | | Deposit date: | 2015-07-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Crystal Structure of the C-Terminal Domain of the Salmonella enterica PduO Protein: An Old Fold with a New Heme-Binding Mode.
Front Microbiol, 7, 2016
|
|
5CV2
 
 | |
2CW0
 
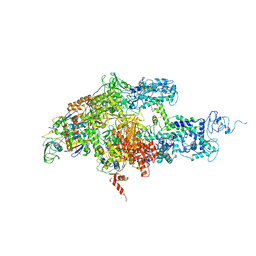 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
2Q6N
 
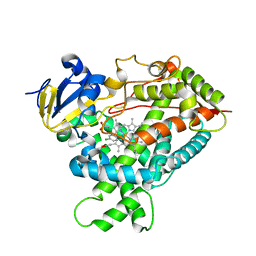 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
5CZ5
 
 | |
5D0W
 
 | | Yeast 20S proteasome beta5-T1S mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5D4D
 
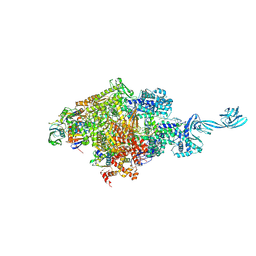 | |
5DJM
 
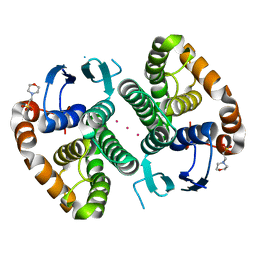 | |
2D1K
 
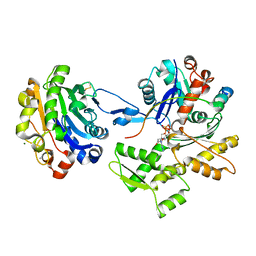 | | Ternary complex of the WH2 domain of mim with actin-dnase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chereau, D, Kerff, F, Dominguez, R. | | Deposit date: | 2005-08-26 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the actin-binding function of missing-in-metastasis
Structure, 15, 2007
|
|
5DNN
 
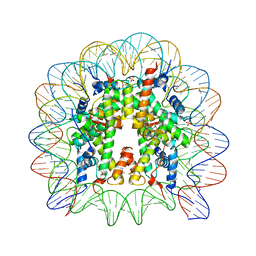 | | Nucleosome core particle containing adducts of gold(I)-triethylphosphane and ruthenium(II)-toluene PTA complexes | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Adhireksan, Z, Ma, Z, Davey, C.A. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric cross-talk in chromatin can mediate drug-drug synergy
Nat Commun, 8, 2017
|
|
5DNM
 
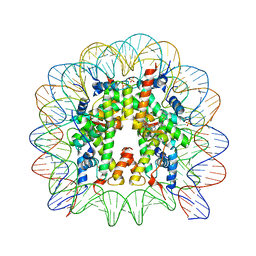 | | Nucleosome core particle containing adducts of ruthenium(II)-toluene PTA complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Adhireksan, Z, Muhammad, R, Davey, C.A. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Allosteric cross-talk in chromatin can mediate drug-drug synergy
Nat Commun, 8, 2017
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2OFR
 
 | |
2C9M
 
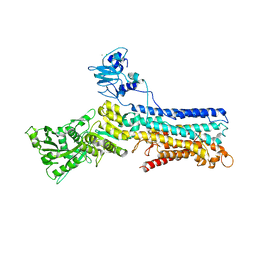 | | Structure of (SR) Calcium-ATPase in the Ca2E1 state solved in a P1 crystal form. | | Descriptor: | CALCIUM ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Lund Jensen, A.-M, Sorensen, T.L.-M, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Modulatory and Catalytic Modes of ATP Binding by the Calcium Pump
Embo J., 25, 2006
|
|
5E78
 
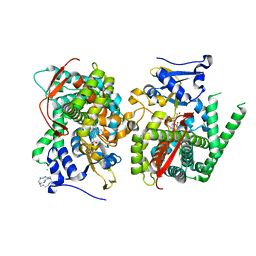 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6CUX
 
 | |
2ESV
 
 | | Structure of the HLA-E-VMAPRTLIL/KK50.4 TCR complex | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Hoare, H.L, Rossjohn, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a major histocompatibility complex class Ib-restricted T cell response
Nat.Immunol., 7, 2006
|
|
2FK3
 
 | |
2FKD
 
 | |
2EWW
 
 | |
6CKR
 
 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | Bromodomain-containing protein 4, N-{3-[2-methyl-6-(1-methyl-1H-pyrazol-4-yl)-1-oxo-1,2-dihydroisoquinolin-4-yl]phenyl}methanesulfonamide | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
