5OJZ
 
 | |
5OK2
 
 | |
5OF2
 
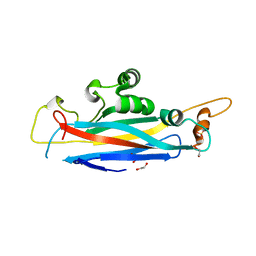 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OK1
 
 | |
3BXU
 
 | | PpcB, A Cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2008-01-14 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the modulation of the redox properties of two Geobacter sulfurreducens homologous triheme cytochromes.
Biochim.Biophys.Acta, 1777, 2008
|
|
5DND
 
 | |
5DNE
 
 | |
5DNC
 
 | |
3BJM
 
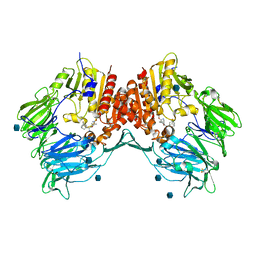 | | Crystal structure of human DPP-IV in complex with (1S,3S, 5S)-2-[(2S)-2-AMINO-2-(3-HYDROXYTRICYCLO[3.3.1.13,7]DEC-1- YL)ACETYL]-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (CAS), (1S,3S,5S)-2-((2S)-2-AMINO-2-(3-HYDROXYADAMANTAN-1- YL)ACETYL)-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (IUPAC), OR BMS-477118 | | Descriptor: | (2~{S})-2-azanyl-1-[(1~{S},3~{S},5~{S})-3-(iminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(5~{R},7~{S})-3-oxidanyl-1-ad amantyl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2007-12-04 | | Release date: | 2008-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Involvement of DPP-IV catalytic residues in enzyme-saxagliptin complex formation.
Protein Sci., 17, 2008
|
|
3F1P
 
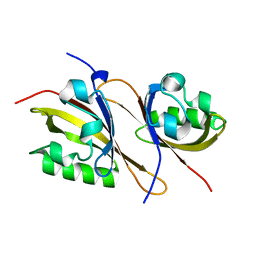 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1O
 
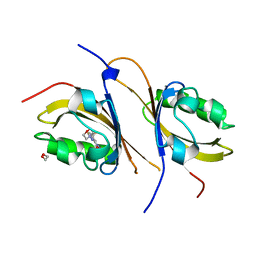 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1N
 
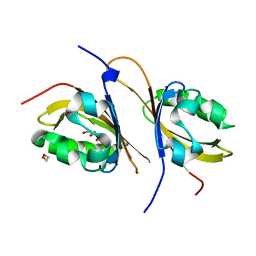 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4P6J
 
 | |
4P6K
 
 | |
4XCP
 
 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
4Y1L
 
 | | Ubc9 Homodimer The Missing Link in Poly-SUMO Chain Formation | | Descriptor: | RWD domain-containing protein 3, SUMO-conjugating enzyme UBC9 | | Authors: | Aileen, Y.A, Ambaye, N.D, Li, Y.J, Vega, R, Bzymek, K, Williams, J.C, Hu, W, Chen, Y. | | Deposit date: | 2015-02-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RWD Domain as an E2 (Ubc9)-Interaction Module.
J.Biol.Chem., 290, 2015
|
|
8PH4
 
 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
4WO1
 
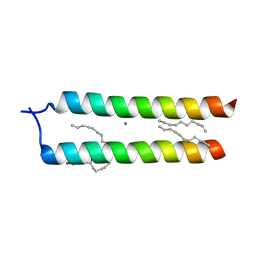 | | Crystal structure of the DAP12 transmembrane domain in lipid cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, TYRO protein tyrosine kinase-binding protein | | Authors: | Call, M.J, Call, M.E, Knoblich, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transmembrane Complexes of DAP12 Crystallized in Lipid Membranes Provide Insights into Control of Oligomerization in Immunoreceptor Assembly.
Cell Rep, 11, 2015
|
|
4LAD
 
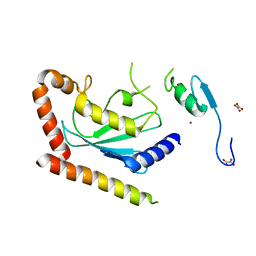 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4YZM
 
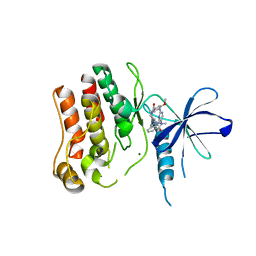 | | Humanized Roco4 bound to LRRK2-In1 | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MAGNESIUM ION, Probable serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Messias, A.C, Ito, G, Sattler, M, Alessi, D.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of LRRK2 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YZN
 
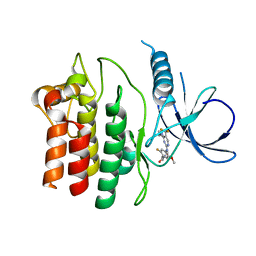 | | Humanized Roco4 bound to Compound 19 | | Descriptor: | (4-{[4-(cyclopropylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-2-fluoro-5-methoxyphenyl)(morpholin-4-yl)methanone, Probable serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Messias, A.C, Ito, G, Sattler, M, Alessi, D.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of LRRK2 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
4WOL
 
 | | Crystal Structure of the DAP12 transmembrane domain in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POTASSIUM ION, TYRO protein tyrosine kinase-binding protein | | Authors: | Call, M.J, Call, M.E, Knoblich, K. | | Deposit date: | 2014-10-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Transmembrane Complexes of DAP12 Crystallized in Lipid Membranes Provide Insights into Control of Oligomerization in Immunoreceptor Assembly.
Cell Rep, 11, 2015
|
|
8HIA
 
 | |
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
