1G1E
 
 | | NMR STRUCTURE OF THE HUMAN MAD1 TRANSREPRESSION DOMAIN SID IN COMPLEX WITH MAMMALIAN SIN3A PAH2 DOMAIN | | Descriptor: | MAD1 PROTEIN, SIN3A | | Authors: | Brubaker, K, Cowley, S.M, Huang, K, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
2JWW
 
 | |
1DRB
 
 | |
2CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-15 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
8SIM
 
 | | KCNQ1 with voltage sensor in the intermediate conformation | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SIN
 
 | | KCNQ1 with voltage sensor in the down conformation | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
1DQS
 
 | | CRYSTAL STRUCTURE OF DEHYDROQUINATE SYNTHASE (DHQS) COMPLEXED WITH CARBAPHOSPHONATE, NAD+ AND ZN2+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (3-DEHYDROQUINATE SYNTHASE), ... | | Authors: | Carpenter, E.P, Hawkins, A.R, Frost, J.W, Brown, K.A. | | Deposit date: | 1998-04-09 | | Release date: | 1999-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis.
Nature, 394, 1998
|
|
3MWU
 
 | | Activated Calcium-Dependent Protein Kinase 1 from Cryptosporidium parvum (CpCDPK1) in complex with bumped kinase inhibitor RM-1-95 | | Descriptor: | 3-(naphthalen-1-ylmethyl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-domain protein kinase 1 | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2010-05-06 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Calcium-Dependent Protein Kinase 1 (CDPK1) from C. parvum and T. gondii.
ACS Med Chem Lett, 1, 2010
|
|
2IO7
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and AMPPNP | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
4O5J
 
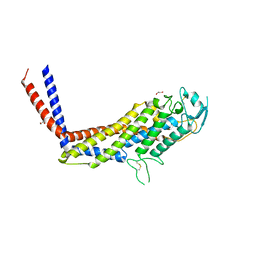 | | Crystal structure of SabA from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Pang, S.S, Nguyen, S.T.S, Whisstock, J.C. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori
J.Biol.Chem., 289, 2013
|
|
1D7A
 
 | | CRYSTAL STRUCTURE OF E. COLI PURE-MONONUCLEOTIDE COMPLEX. | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, PHOSPHORIBOSYLAMINOIMIDAZOLE CARBOXYLASE | | Authors: | Mathews, I.I, Kappock, T.J, Stubbe, J, Ealick, S.E. | | Deposit date: | 1999-10-16 | | Release date: | 1999-12-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway.
Structure Fold.Des., 7, 1999
|
|
8T8F
 
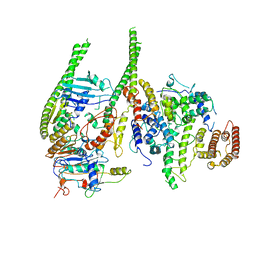 | | Smc5/6 8mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosome element 5, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8T8E
 
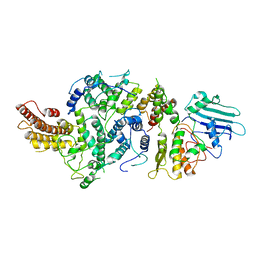 | | cryoEM structure of Smc5/6 5mer | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 6 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2XN4
 
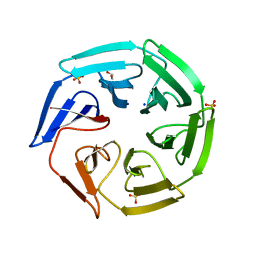 | | Crystal structure of the kelch domain of human KLHL2 (Mayven) | | Descriptor: | 1,2-ETHANEDIOL, KELCH-LIKE PROTEIN 2, SODIUM ION, ... | | Authors: | Canning, P, Hozjan, V, Cooper, C.D.O, Ayinampudi, V, Vollmar, M, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
7BVY
 
 | |
2YB0
 
 | | The Crystal Structure of Leishmania major dUTPase in complex deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, DUTPASE, SULFATE ION | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
8G2J
 
 | | Hybrid aspen cellulose synthase-8 bound to UDP-glucose | | Descriptor: | Cellulose synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
8G27
 
 | | Hybrid aspen cellulose synthase-8 bound to UDP | | Descriptor: | Cellulose synthase, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-03-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
7RHR
 
 | | Cryo-EM structure of Xenopus Patched-1 in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Huang, P, Lian, T, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
2YPF
 
 | | Structure of the AvrBs3-DNA complex provides new insights into the initial thymine-recognition mechanism | | Descriptor: | 5'-D(*TP*AP*GP*AP*GP*GP*GP*TP*TP*AP*GP*GP*TP*TP *TP*AP*TP*AP*TP*AP*AP)-3', 5'-D(*TP*TP*TP*AP*TP*AP*TP*AP*AP*AP*CP*CP*TP*AP *AP*CP*CP*CP*TP*CP*TP*AP)-3', AVRBS3 | | Authors: | Stella, S, Molina, R, Yefimenko, I, Prieto, J, Silva, G.H, Bertonati, C, Juillerat, A, Duchateau, P, Montoya, G. | | Deposit date: | 2012-10-30 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Avrbs3-DNA Complex Provides New Insights Into the Initial Thymine-Recognition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7RHQ
 
 | | Cryo-EM structure of Xenopus Patched-1 in complex with GAS1 and Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, P, Lian, T, Wierbowski, B, Garcia-Linares, S, Jiang, J, Salic, A. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for catalyzed assembly of the Sonic hedgehog-Patched1 signaling complex.
Dev.Cell, 57, 2022
|
|
6BY4
 
 | | Single-State 14-mer UUCG Tetraloop calculated from Exact NOEs | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
2ADR
 
 | |
6BY5
 
 | | Two-State 14-mer UUCG Tetraloop calculated from Exact NOEs (State one: Conformers 1-5, State Two: Conformers 6-10) | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
