4WHU
 
 | | BROMO domain of CREB binding protein | | Descriptor: | 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, CREB-binding protein | | Authors: | Liu, S. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
4WHW
 
 | | Direct photocapture of bromodomains using tropolone chemical probes | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, Bromodomain-containing protein 4 | | Authors: | Hett, E.C, Piatnitski Chekler, E.L, Basak, A, Bonin, P.D, Denny, R.A, Flick, A.C, Geoghegan, K.F, Liu, S, Pletcher, M.T, Robinson, R.P, Sahasrabudhe, P, Salter, S, Stock, I.A, Jones, L.H. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
8HOE
 
 | |
4WUK
 
 | | Crystal structure of apo CH65 Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CH65 heavy chain, CH65 light chain | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2014-11-01 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the apo anti-influenza CH65 Fab.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8HOC
 
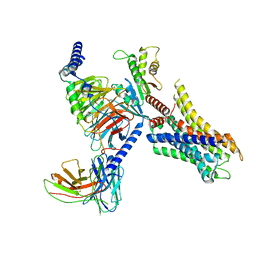 | | Cryo-EM structure of ligand histamine-bound Histamine H4 receptor Gi complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tang, W.Q, Sun, X.Y, Li, F.H, Wang, J.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
To Be Published
|
|
4WKH
 
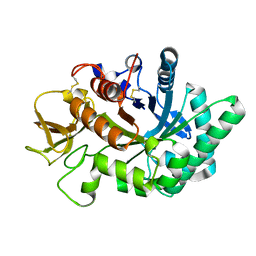 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8HQB
 
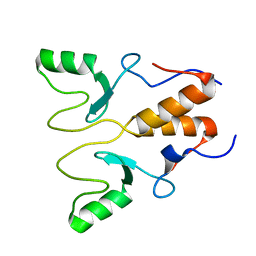 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
4WKQ
 
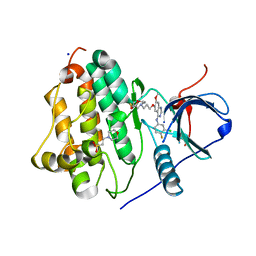 | |
8HNQ
 
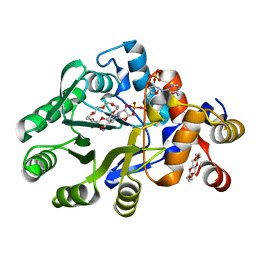 | | The structure of a alcohol dehydrogenase AKR13B2 with NADP | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of AKR13B2
To Be Published
|
|
8HPF
 
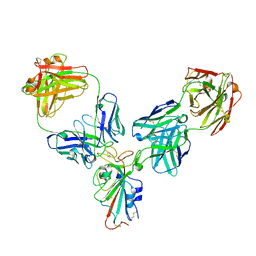 | |
4WKV
 
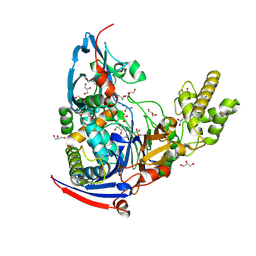 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, trihydroxy(octyl)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1434 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
8HQV
 
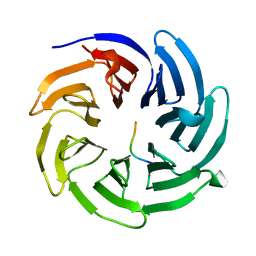 | |
4WUU
 
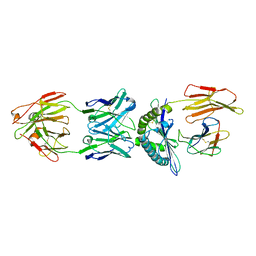 | | Structure of ESK1 in complex with HLA-A*0201/WT1 | | Descriptor: | ARG-MET-PHE-PRO-ASN-ALA-PRO-TYR-LEU, Beta-2-microglobulin, ESK1, ... | | Authors: | Ataie, N.J, Ng, H.L. | | Deposit date: | 2014-11-03 | | Release date: | 2015-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Structure of a TCR-Mimic Antibody with Target Predicts Pharmacogenetics.
J.Mol.Biol., 428, 2016
|
|
4WVA
 
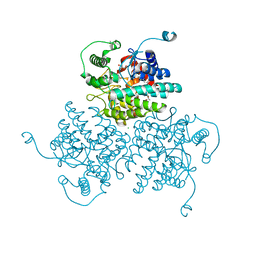 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
8HPP
 
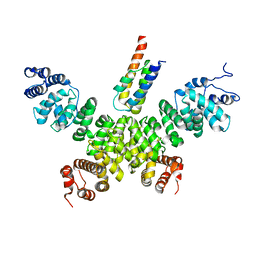 | | Crystal structure of human INTS3 with SAGE1 | | Descriptor: | Integrator complex subunit 3, Sarcoma antigen 1 | | Authors: | Deng, W, Wu, J, Lei, M. | | Deposit date: | 2022-12-12 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cancer-testis antigen SAGE1 is a pan-cancer master regulator of RNA polymerase II
To Be Published
|
|
8HRL
 
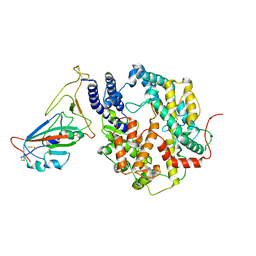 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRJ
 
 | |
4WLE
 
 | |
4WLS
 
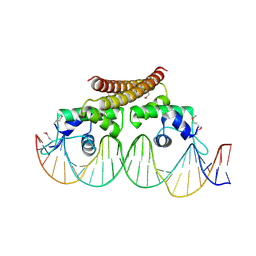 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
4WVG
 
 | |
4WVT
 
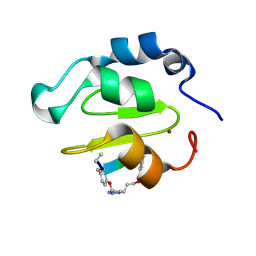 | | Crystal structure of XIAP-BIR2 domain complexed with ligand bound | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
4WW5
 
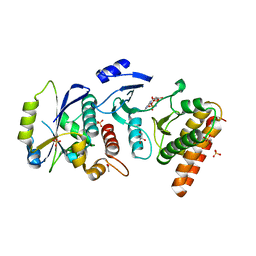 | | Crystal structure of binary complex Bud32-Cgi121 in complex with AMPP | | Descriptor: | ACETATE ION, EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-11-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
4WM0
 
 | |
4WME
 
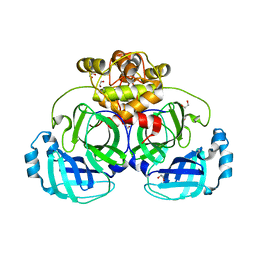 | | Crystal structure of catalytically inactive MERS-CoV 3CL Protease (C148A) in spacegroup C2 | | Descriptor: | 1,2-ETHANEDIOL, MERS-CoV 3CL protease | | Authors: | Lountos, G.T, Needle, D, Waugh, D.S. | | Deposit date: | 2014-10-08 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WN2
 
 | |
