4AQY
 
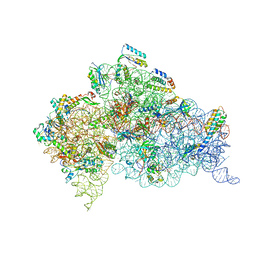 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFL
 
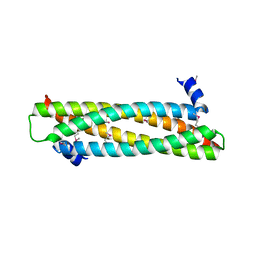 | | The crystal structure of the ING4 dimerization domain reveals the functional organization of the ING family of chromatin binding proteins. | | Descriptor: | INHIBITOR OF GROWTH PROTEIN 4 | | Authors: | Culurgioni, S, Munoz, I.G, Moreno, A, Palacios, A, Villate, M, Palmero, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-22 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystal Structure of Inhibitor of Growth 4 (Ing4) Dimerization Domain Reveals Functional Organization of Ing Family of Chromatin-Binding Proteins.
J.Biol.Chem., 287, 2012
|
|
2RHB
 
 | | Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Palaninathan, S, Bhardwaj, K, Alcantara, J.M.O, Guarino, L, Yi, L.L, Kao, C.C, Sacchettini, J. | | Deposit date: | 2007-10-08 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.
J.Biol.Chem., 283, 2008
|
|
5NPW
 
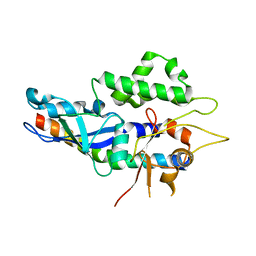 | | Structure of human ATG5-ATG16L1(ATG5BD) complex (C2) | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Archna, A, Scrima, A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification, biochemical characterization and crystallization of the central region of human ATG16L1.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6U72
 
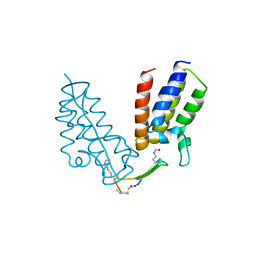 | | BRD4-BD1 in complex with the cyclic peptide 3.1_2_AcK5toA | | Descriptor: | 3.1_2_AcK5toA, AMINO GROUP, Bromodomain-containing protein 4 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P, Mouradian, K.S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NSR
 
 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
5NWY
 
 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
5ZGH
 
 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
5ZGB
 
 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-08 | | Release date: | 2018-04-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NDG
 
 | | Crystal structure of geneticin (G418) bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NSS
 
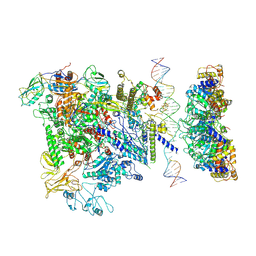 | | Cryo-EM structure of RNA polymerase-sigma54 holoenzyme with promoter DNA and transcription activator PspF intermedate complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta',DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
6UH5
 
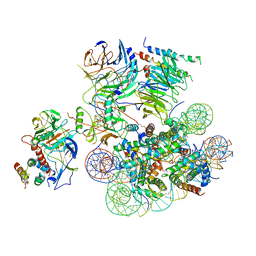 | | Structural basis of COMPASS eCM recognition of the H2Bub nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
4ADX
 
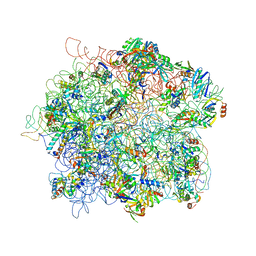 | | The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 | | Descriptor: | 23S Ribosomal RNA EXPANSION SEGMENTS, 23S ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Godinic-Mikulcic, V, Crnkovic, A, Ibba, M, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-Em Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution
J.Mol.Biol., 418, 2012
|
|
6ULQ
 
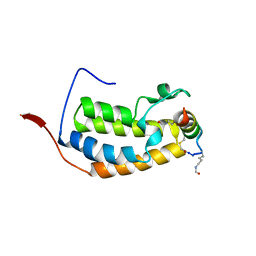 | | BRD2-BD1 in complex with the cyclic peptide 4.2_3 | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 4.2_3 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-10-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UKI
 
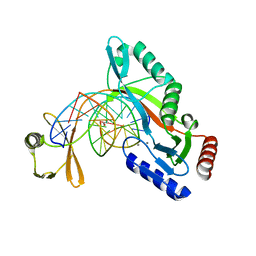 | |
2V0W
 
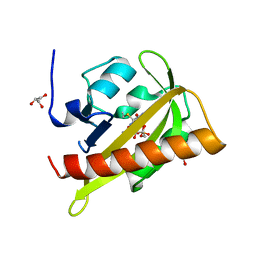 | |
4B2D
 
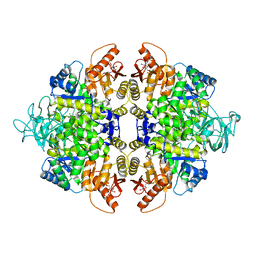 | | human PKM2 with L-serine and FBP bound. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVATE KINASE ISOZYMES M1/M2, ... | | Authors: | Chaneton, B, Hillmann, P, Zheng, L, Martin, A.C.L, Maddocks, O.D.K, Chokkathukalam, A, Coyle, J.E, Jankevics, A, Holding, F.P, Vousden, K.H, Frezza, C, O'Reilly, M, Gottlieb, E. | | Deposit date: | 2012-07-13 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serine is a natural ligand and allosteric activator of pyruvate kinase M2.
Nature, 491, 2012
|
|
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Bai, R, Wan, R, Yan, C, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
6ULW
 
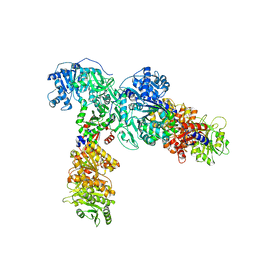 | | Adenylation, ketoreductase, and pseudo Asub multidomain structure of a keto acid-selecting NRPS module | | Descriptor: | Amino acid adenylation domain-containing protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Alonzo, D.A, Wang, J, Chiche-Lapierre, C, Schmeing, T.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
2CGT
 
 | | GROEL-ADP-gp31 COMPLEX | | Descriptor: | 60 KDA GROEL, CAPSID ASSEMBLY PROTEIN GP31 | | Authors: | Clare, D.K, Bakkes, P.J, van Heerikhuizen, H, van der Vies, S.M, Saibil, H.R. | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | An Expanded Protein Folding Cage in the Groel-Gp31 Complex.
J.Mol.Biol., 358, 2006
|
|
6SV4
 
 | | The cryo-EM structure of SDD1-stalled collided trisome. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-09-17 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2C7E
 
 | | REVISED ATOMIC STRUCTURE FITTING INTO A GROEL(D398A)-ATP7 CRYO-EM MAP (EMD 1047) | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-02-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy
Cell(Cambridge,Mass.), 107, 2001
|
|
6SW9
 
 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
2C7D
 
 | | Fitted coordinates for GroEL-ADP7-GroES Cryo-EM complex (EMD-1181) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
