6I44
 
 | | Allosteric activation of human prekallikrein by apple domain disc rotation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Pathak, M, MaCrae, K, Dreveny, I, Emsley, J. | | Deposit date: | 2018-11-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J.Thromb.Haemost., 17, 2019
|
|
7AVM
 
 | | Crystal Structure of Pro-Rhodesain C150A | | Descriptor: | Cysteine protease, GLYCEROL | | Authors: | Johe, P, Jaenicke, E, Neuweiler, H, Schirmeister, T, Kersten, C, Hellmich, U. | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, interdomain dynamics, and pH-dependent autoactivation of pro-rhodesain, the main lysosomal cysteine protease from African trypanosomes.
J.Biol.Chem., 296, 2021
|
|
7A8X
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with the HMA domain of Pikh-1 from rice (Oryza sativa) | | Descriptor: | AVR-Pik protein, NBS-LRR class disease resistance protein | | Authors: | Maidment, J.H.R, Xiao, G, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
3DTX
 
 | | Crystal structure of HLA-B*2705 complexed with the double citrullinated vasoactive intestinal peptide type 1 receptor (VIPR) peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, Double citrullinated vasoactive intestinal polypeptide receptor, MHC class I antigen (Fragment) | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2008-07-16 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Citrullination-and mhc polymorphism-dependent conformational changes of a self peptide
To be Published
|
|
5BWY
 
 | | Structure of proplasmepsin II from Plasmodium falciparum, Space Group P43212 | | Descriptor: | Plasmepsin-2 | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-06-08 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5FFN
 
 | | Complex of subtilase SubTY from Bacillus sp. TY145 with chymotrypsin inhibitor CI2A | | Descriptor: | CALCIUM ION, Enzyme subtilase SubTY from Bacillus sp. TY145, SODIUM ION, ... | | Authors: | McAuley, K.E, Svendsen, A, Oestergaard, P.R, Dohnalek, J, Wilson, K.S. | | Deposit date: | 2015-12-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
4AXM
 
 | | TRIAZINE CATHEPSIN INHIBITOR COMPLEX | | Descriptor: | 4-[(4-chlorobenzyl)(cyclohexyl)amino]-6-morpholin-4-yl-1,3,5-triazine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Ehmke, V, Diederich, F, Banner, D.W, Benz, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Triazine Nitriles as Rhodesain Inhibitors: Structure-Activity Relationships, Bioisosteric Imidazopyridine Nitriles, and X-Ray Crystal Structure Analysis with Human Cathepsin L
Chemmedchem, 8, 2013
|
|
4B5R
 
 | | SAM-I riboswitch bearing the H. marismortui K-t-7 | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Daldrop, P, Lilley, D.M.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Plasticity of a Structural Motif in RNA: Structural Polymorphism of a Kink Turn as a Function of its Environment.
RNA, 19, 2013
|
|
4AXL
 
 | | HUMAN CATHEPSIN L APO FORM WITH ZN | | Descriptor: | ACETATE ION, CATHEPSIN L1, GLYCEROL, ... | | Authors: | Banner, D.W, Benz, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Optimization of Triazine Nitriles as Rhodesain Inhibitors: Structure-Activity Relationships, Bioisosteric Imidazopyridine Nitriles, and X-Ray Crystal Structure Analysis with Human Cathepsin L
Chemmedchem, 8, 2013
|
|
4BG6
 
 | | 14-3-3 interaction with Rnd3 prenyl-phosphorylation motif | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FARNESYL, ... | | Authors: | Riou, P, Kjaer, S, Purkiss, A, O'Reilly, N, McDonald, N.Q. | | Deposit date: | 2013-03-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 Proteins Interact with a Hybrid Prenyl-Phosphorylation Motif to Inhibit G Proteins.
Cell(Cambridge,Mass.), 153, 2013
|
|
6I58
 
 | | Allosteric activation of human prekallikrein by apple domain disc rotation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, C, Pathak, M, McCrae, K, Dreveny, I, Emsley, J. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J.Thromb.Haemost., 17, 2019
|
|
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
1KXC
 
 | |
1KXA
 
 | |
1KXD
 
 | |
1KXB
 
 | |
1KXE
 
 | |
1KXF
 
 | |
4H6T
 
 | | Crystal structure of prethrombin-2 mutant E14eA/D14lA/E18A/S195A | | Descriptor: | PHOSPHATE ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
2M3S
 
 | | Calmodulin, i85l, f92e, h107i, l112r, a128t, m144r mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Moroz, Y.S, Wu, Y, Cheng, H, Roder, H, Korendovych, I.V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A single mutation in a regulatory protein produces evolvable allosterically regulated catalyst of nonnatural reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6ASC
 
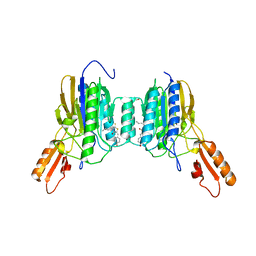 | | Mre11 dimer in complex with Endonuclease inhibitor PFM04 | | Descriptor: | (5E)-3-butyl-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, MANGANESE (II) ION, ... | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Allostery with Avatars to Design Inhibitors Assessed by Cell Activity: Dissecting MRE11 Endo- and Exonuclease Activities.
Meth. Enzymol., 601, 2018
|
|
4YV3
 
 | |
4YPC
 
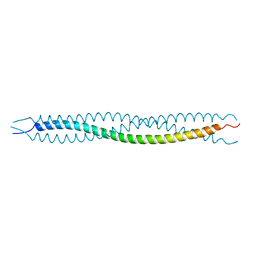 | |
7U63
 
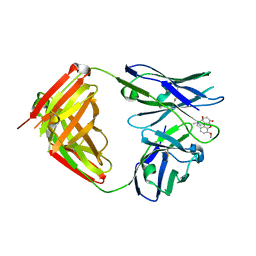 | | Crystal Structure Anti-Oxycodone Antibody HY2-A12 Fab Complexed with Oxycodone | | Descriptor: | 14-hydroxy-3-methoxy-17-methyl-5beta-4,5-epoxymorphinan-6-one, HY2-A12 Fab Heavy Chain, HY2-A12 Fab Light Chain | | Authors: | Rodarte, J.V, Pancera, M.P, Weidle, C, Rupert, P.B, Strong, R.K. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
2BBX
 
 | |
