5C2F
 
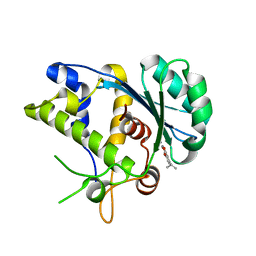 | | K428A mutant nuclease domain of the large terminase subunit gp2 of bacterial virus Sf6 with Manganese and beta-thujaplicinol | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, Gene 2 protein, MANGANESE (II) ION | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Two distinct modes of metal ion binding in the nuclease active site of a viral DNA-packaging terminase: insight into the two-metal-ion catalytic mechanism.
Nucleic Acids Res., 43, 2015
|
|
2J85
 
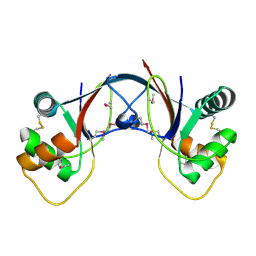 | |
2IG0
 
 | | Structure of 53BP1/methylated histone peptide complex | | Descriptor: | Dimethylated Histone H4-K20 peptide, SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Mer, G. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
7NYY
 
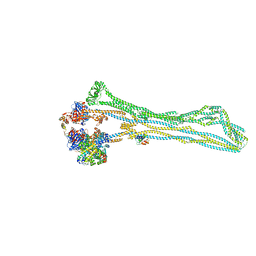 | | Cryo-EM structure of the MukBEF monomer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
7NZ4
 
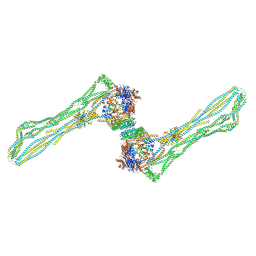 | | Cryo-EM structure of the MukBEF dimer | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | Authors: | Buermann, F, Lowe, J. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
2JPB
 
 | |
7TUJ
 
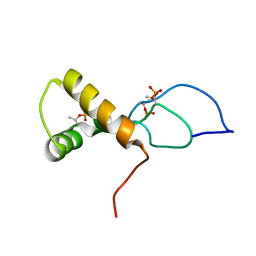 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
5CR5
 
 | |
6FXA
 
 | |
6KDN
 
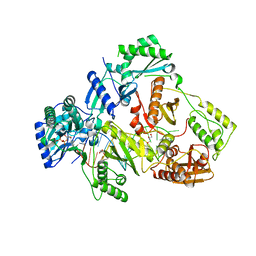 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
5C10
 
 | |
5C2D
 
 | |
453D
 
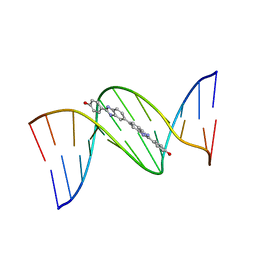 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'-BENZIMIDAZOLE COMPLEX | | Descriptor: | 4-{[4-HYDROXY-PHENYL]-1H-BENZIMIDAZOLE-5-YL}-BENZIMIDAZOLE-2-YL-[4-HYDROXY-BENZENE], DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Neidle, S. | | Deposit date: | 1999-02-18 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric bis-benzimidazoles, a new class of sequence-selective DNA-binding molecules
J.Chem.Soc.,Chem.Commun., 1999
|
|
6IFO
 
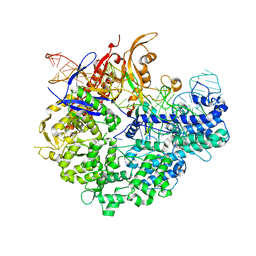 | | Crystal structure of AcrIIA2-SpyCas9-sgRNA ternary complex | | Descriptor: | AcrIIA2, CRISPR-associated endonuclease Cas9/Csn1, RNA (99-MER) | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race.
Mol. Cell, 73, 2019
|
|
2KUO
 
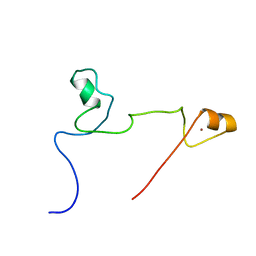 | | Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Li, G.Y, McCulloch, R.D, Fenton, A, Cheung, M, Meng, L, Ikura, M, Koch, C.A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and identification of ADP-ribose recognition motifs of aprataxin PNK-like factor (APLF) required for the interaction with sites of DNA damage response
To be Published
|
|
5C15
 
 | |
6KDK
 
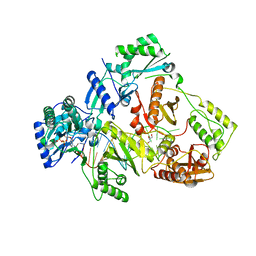 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
7NPD
 
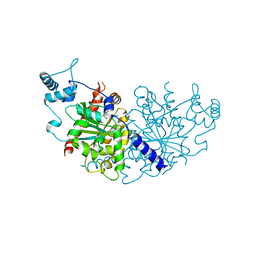 | | Vibiro cholerae ParA2 | | Descriptor: | Walker A-type ATPase | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
7Z4J
 
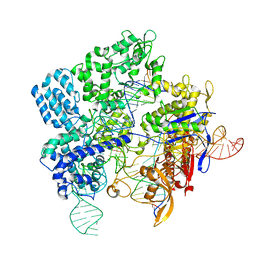 | |
7YMO
 
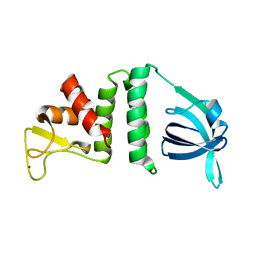 | |
3VA3
 
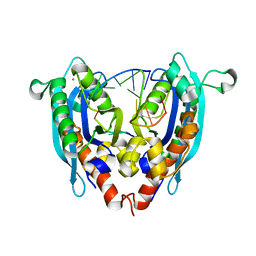 | |
6IK9
 
 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
3HG8
 
 | |
8CIL
 
 | |
1WTU
 
 | | TRANSCRIPTION FACTOR 1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSCRIPTION FACTOR 1 | | Authors: | Jia, X, Grove, A, Ivancic, M, Hsu, V.L, Geiduschek, E.P, Kearns, D.R. | | Deposit date: | 1996-07-29 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution.
J.Mol.Biol., 263, 1996
|
|
