2UWX
 
 | | Active site restructuring regulates ligand recognition in class A penicillin-binding proteins | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, DiGuilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6RG4
 
 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzyl-4-methylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(3~{S})-4-methyl-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6DOR
 
 | |
9D02
 
 | |
5XZF
 
 | | Vitamin D receptor with a synthetic ligand ADRO1 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6S)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
6OPP
 
 | | Asymmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with DDM | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
5SY8
 
 | |
5ZJD
 
 | | Lactate dehydrogenase with NADH and MLA | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
5ZOI
 
 | | Crystal Structure of alpha1,3-Fucosyltransferase | | Descriptor: | Alpha-(1,3)-fucosyltransferase FucT, [[(2S,3R,4S,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5R,6R)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Tan, Y, Yang, G. | | Deposit date: | 2018-04-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Directed evolution of an alpha 1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method.
Sci Adv, 5, 2019
|
|
7MN0
 
 | |
9GGP
 
 | | Alpha-1-antitrypsin in complex with the Fab fragment of an anti-polymer antibody | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1-antitrypsin, Fab fragment heavy chain of 2C1 monoclonal antibody, ... | | Authors: | Lowen, S.M, Laffranchi, M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-resolution characterization of ex vivo AAT polymers by solution-state NMR spectroscopy.
Sci Adv, 11, 2025
|
|
5HIK
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
6RX6
 
 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
8ITV
 
 | | KL2.1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (3~{S},5~{R},6~{E},8~{Z},10~{R},12~{E},14~{E},16~{S})-3,16-bis(azanyl)-8,10,12-trimethyl-16-[(2~{S},4~{R},5~{S},6~{S})-5-methyl-4-oxidanyl-6-[(~{E})-prop-1-enyl]oxan-2-yl]-5-oxidanyl-hexadeca-6,8,12,14-tetraenoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KL2.1 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
5H9O
 
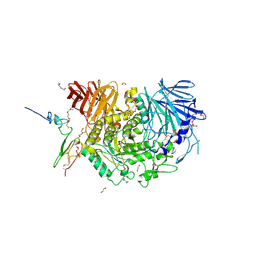 | | Complex of Murine endoplasmic reticulum alpha-glucosidase II with D-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-12-29 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8WMF
 
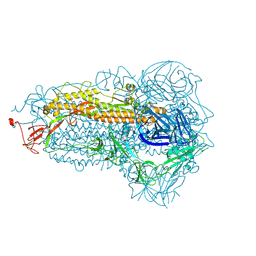 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
9MZY
 
 | | Crystal structure of human RIPK1 with Compound 22 | | Descriptor: | 1-[(2S,5S)-2,3-dihydro-2,5-methano-1,4-benzoxazepin-4(5H)-yl]-3,3-difluoro-2,2-dimethylpropan-1-one, CHLORIDE ION, GLYCINE, ... | | Authors: | Palte, R.L, Lesburg, C.A, Maskos, K, Thomsen, M, Lammens, A. | | Deposit date: | 2025-01-23 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Discovery of Bridged Benzoazepine Amides as Selective Allosteric Modulators of RIPK1.
Acs Med.Chem.Lett., 16, 2025
|
|
9N36
 
 | | CryoEM structure Of Respiratory Syncytial Virus Polymerase with novel non-nucleoside inhibitor compound 22 | | Descriptor: | 5-(5-bromo-1-methyl-2-oxo-1,2-dihydrospiro[indole-3,4'-piperidin]-1'-yl)-3-chloropyridine-2-carbonitrile, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yin, Y, Yu, X, Kalin, J.H, Tran, M.T, Sharma, S. | | Deposit date: | 2025-01-30 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | CryoEM structure Of Respiratory Syncytial Virus Polymerase in complex with novel non-nucleoside inhibitor 5-(5-bromo-1-methyl-2-oxo-1,2-dihydrospiro[indole-3,4'-piperidin]-1'-yl)-3-chloropyridine-2-carbonitrile (compound 22)
To Be Published
|
|
5FKJ
 
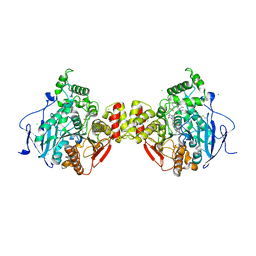 | | Crystal structure of mouse acetylcholinesterase in complex with C-547, an alkyl ammonium derivative of 6-methyl uracil | | Descriptor: | 1,3-BIS[5(DIETHYL-O-NITROBENZYLAMMONIUM)PENTYL]-6-METHYLURACIL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Nachon, F, Villard-Wandhammer, M, Petrov, K, Masson, P. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Slow-Binding Inhibition of Acetylcholinesterase by a 6-Methyluracil Alkyl-Ammonium Derivative: Mechanism and Advantages for Myasthenia Gravis Treatment.
Biochem.J., 473, 2016
|
|
6I48
 
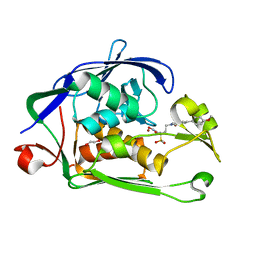 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6TZJ
 
 | | ADC-7 in complex with boronic acid transition state inhibitor ME_096 | | Descriptor: | Beta-lactamase, GLYCINE, [4-[[(4-methylphenyl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]methyl-phosphonooxy-borinic acid | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
6ORN
 
 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch bNAb 10-1074 | | Descriptor: | 10-1074 antibody Fab heavy chain, 10-1074 antibody Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
3MAU
 
 | | Crystal structure of StSPL in complex with phosphoethanolamine | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, sphingosine-1-phosphate lyase, ... | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-24 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
6XOX
 
 | | cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970 | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, Alpha subunit of Gs with N-terminus swapped with equivalent residues in Gi,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Kobilka, B.K, Sloop, K.W, Feng, D, Kobilka, T.S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RDM
 
 | |
