2UXD
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
6TUT
 
 | | Cryo-EM structure of the RNA Polymerase III-Maf1 complex | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Vorlaender, M.K, Hagen, W.J.H, Mueller, C.W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for RNA polymerase III transcription repression by Maf1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7UND
 
 | | Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex (stalled at +38) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Filipovski, M, Vos, S.M, Farnung, L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosome retention during transcription elongation.
Science, 376, 2022
|
|
6X67
 
 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | Descriptor: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | Authors: | Chen, Q, Hickman, A.B, Dyda, F. | | Deposit date: | 2020-05-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6XBY
 
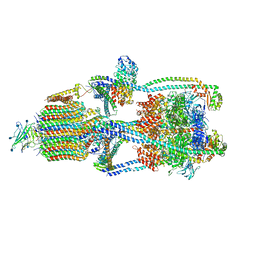 | | Cryo-EM structure of V-ATPase from bovine brain, state 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
6U47
 
 | |
2UXB
 
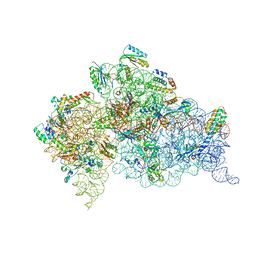 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
6C46
 
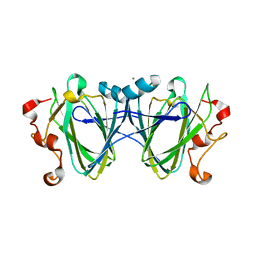 | |
6C5L
 
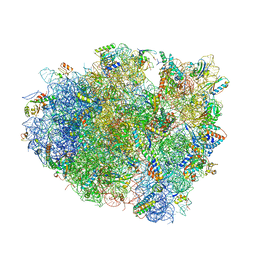 | |
7TOS
 
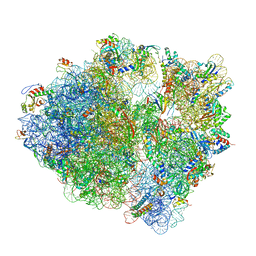 | | E. coli 70S ribosomes bound with the ALS/FTD-associated dipeptide repeat protein PR20 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
5DOY
 
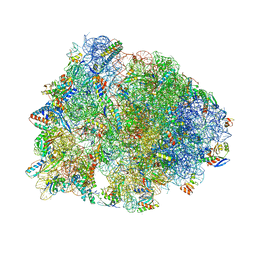 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic Hygromycin A, mRNA and three tRNAs in the A, P and E sites at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5MQV
 
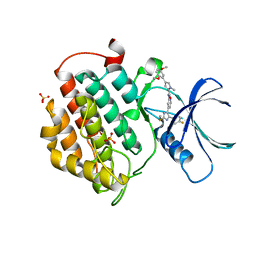 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
6XBW
 
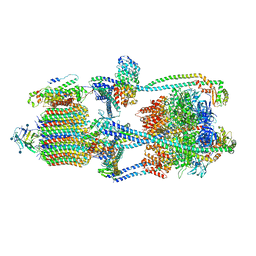 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
8R08
 
 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
5FJ9
 
 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6XHV
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6EMY
 
 | | Structure of the Tn1549 transposon Integrase (aa 82-397, Y379F) in complex with transposon right end DNA | | Descriptor: | DNA (20-MER), DNA (26-MER), Int protein | | Authors: | Schulz, E.C, Rubio-Cosials, A, Barabas, O. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance.
Cell, 173, 2018
|
|
5F99
 
 | |
2VJU
 
 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
5NDW
 
 | | Crystal structure of aminoglycoside TC007 bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7V2R
 
 | | Active state complex I from Q1-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2D
 
 | | Deactive state complex I from Q10 dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-08 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V33
 
 | | Active state complex I from rotenone-NADH dataset | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7V2K
 
 | | Deactive state complex I from DQ-NADH dataset | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
