7OK9
 
 | |
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
7O4A
 
 | |
7O49
 
 | |
5OAU
 
 | |
2ZC3
 
 | | Penicillin-binding protein 2X (PBP 2X) acyl-enzyme complex (biapenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(6,7-dihydro-5H-pyrazolo[1,2-a][1,2,4]triazol-4-ium-6-ylsulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-meth yl-4,5-dihydro-1H-pyrrole-2-carboxylate, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
7O4B
 
 | |
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
5UY7
 
 | |
4KQO
 
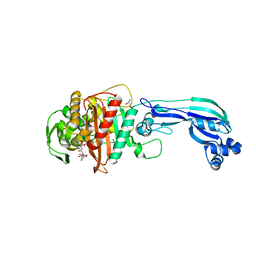 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
1VQQ
 
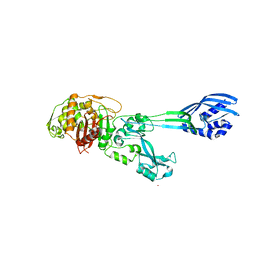 | |
6TUD
 
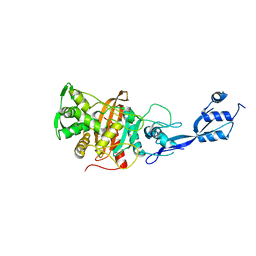 | |
4CPK
 
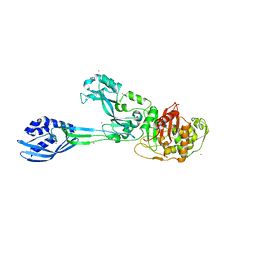 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
3VSK
 
 | |
3VSL
 
 | | Crystal structure of penicillin-binding protein 3 (PBP3) from methicilin-resistant Staphylococcus aureus in the cefotaxime bound form. | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Yoshida, H, Tame, J.R, Park, S.Y. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Penicillin-Binding Protein 3 (PBP3) from Methicillin-Resistant Staphylococcus aureus in the Apo and Cefotaxime-Bound Forms.
J.Mol.Biol., 423, 2012
|
|
6VOT
 
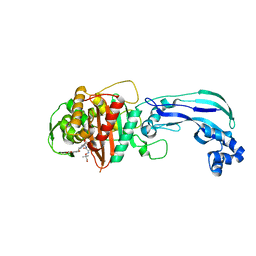 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | Descriptor: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F. | | Deposit date: | 2020-01-31 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
6G88
 
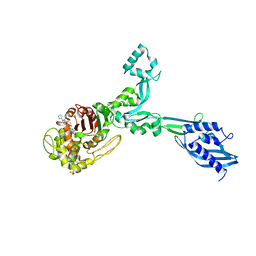 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6G9F
 
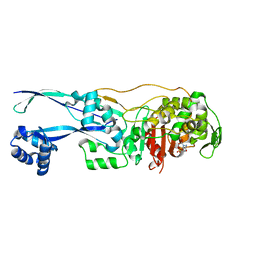 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G0K
 
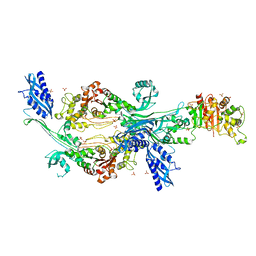 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6VJE
 
 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
6G9S
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | (3~{R},6~{S})-6-(aminomethyl)-4-(1,3-oxazol-5-yl)-3-(sulfooxyamino)-3,6-dihydro-2~{H}-pyridine-1-carboxylic acid, Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
6G9P
 
 | | Structural basis for the inhibition of E. coli PBP2 | | Descriptor: | Peptidoglycan D,D-transpeptidase MrdA | | Authors: | Ruff, M, Levy, N. | | Deposit date: | 2018-04-11 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural Basis for E. coli Penicillin Binding Protein (PBP) 2 Inhibition, a Platform for Drug Design.
J.Med.Chem., 62, 2019
|
|
