8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8RS0
 
 | |
8RRX
 
 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RRW
 
 | |
8RRV
 
 | |
8RRU
 
 | |
8RRT
 
 | |
8R5K
 
 | |
8QMO
 
 | | Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Kwong, H.S, Grandvuillemin, L, Sirounian, S, Ancelin, A, Lai-Kee-Him, J, Carivenc, C, Lancey, C, Ragan, T.J, Hesketh, E.L, Bourguet, W, Gruszczyk, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
J.Mol.Biol., 436, 2024
|
|
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
8POD
 
 | | Crystal structure of the kinase domain of ACVR1 (ALK2) in complex with FKBP12 and MU1700 | | Descriptor: | 6-(4-piperazin-1-ylphenyl)-3-quinolin-4-yl-furo[3,2-b]pyridine, Activin receptor type-1, FLUORIDE ION, ... | | Authors: | Cros, J, Baltanas-Copado, J, Knapp, S, Paruch, K, Nemec, N, Bullock, A.N. | | Deposit date: | 2023-07-04 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of the kinase domain of ACVR1 (ALK2) in complex with FKBP12 and MU1700
To Be Published
|
|
8PJA
 
 | | FKBP51FK1 F67E/K58 (i, i+9) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
8PJ8
 
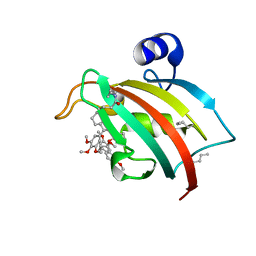 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
8PDF
 
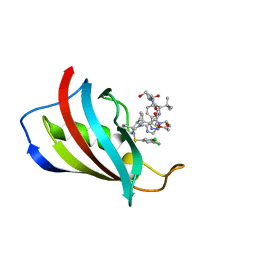 | | FKBP12 in complex with PROTAC 6a2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[4-[(1~{S})-1-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PC2
 
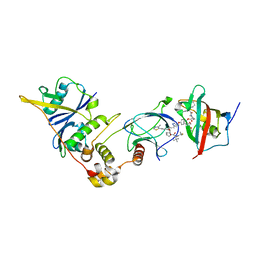 | | SelDeg51 in complex with FKBP51FK1 domain and pVHL:EloB:EloC | | Descriptor: | Elongin-B, Elongin-C, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8P42
 
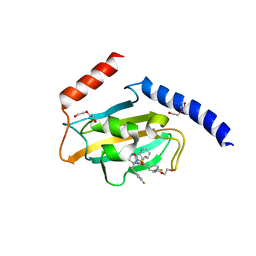 | | Full length structure of TcMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Macrophage infectivity potentiator | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-19 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P3D
 
 | | Full length structure of TcMIP with bound inhibitor NJS224. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-(pyridin-3-ylmethylamino)pentan-2-yl]piperidine-2-carboxamide, SODIUM ION, peptidylprolyl isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P3C
 
 | | Full length structure of BpMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8JGA
 
 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8JD4
 
 | | Cryo-EM structure of G protein-free mGlu2-mGlu4 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
