3JC7
 
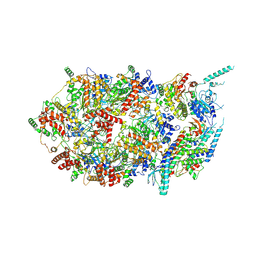 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1K6J
 
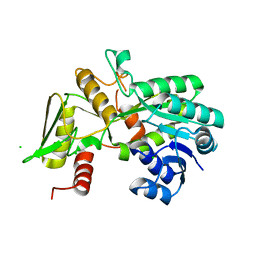 | | Crystal structure of Nmra, a negative transcriptional regulator (Monoclinic form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
5T1J
 
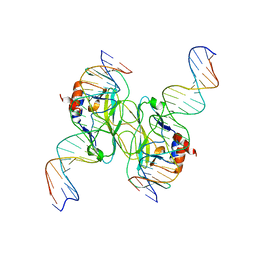 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5THV
 
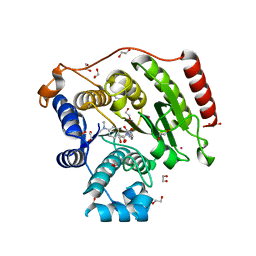 | | Crystal Structure of G305A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5N9Q
 
 | | Structure of A. thaliana RCD1(468-567) | | Descriptor: | Inactive poly [ADP-ribose] polymerase RCD1 | | Authors: | Tossavainen, H, Hellman, M, Vainonen, J, Kangasjarvi, J, Permi, P. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors.
Elife, 8, 2019
|
|
5THT
 
 | | Crystal Structure of G303A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5VK3
 
 | | Apo ctPRC2 with E840A and K852D mutations in Ezh2 | | Descriptor: | Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12, Polycomb Protein EED, ZINC ION | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-04-20 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
3JC5
 
 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5THS
 
 | | Crystal Structure of G302A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5THU
 
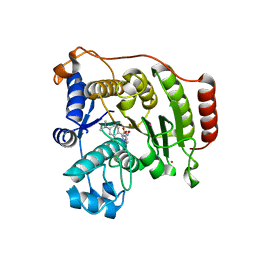 | | Crystal Structure of G304A HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
3JC6
 
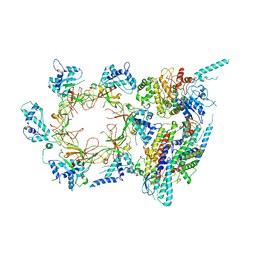 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1K6X
 
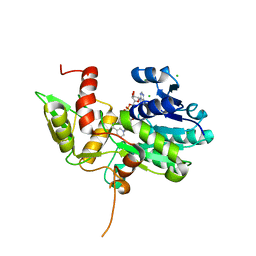 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
2NNY
 
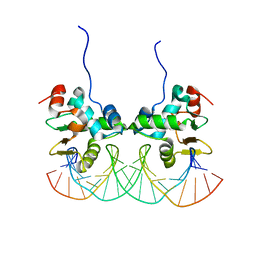 | | Crystal structure of the Ets1 dimer DNA complex. | | Descriptor: | 5'-D(*A*CP*TP*CP*CP*AP*GP*GP*AP*AP*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*TP*CP*T)-3', 5'-D(*T*AP*GP*AP*CP*AP*GP*GP*AP*AP*GP*CP*AP*CP*TP*TP*CP*CP*TP*GP*GP*AP*G)-3', C-ets-1 protein | | Authors: | Lamber, E.P, Kachalova, G.S, Wilmanns, M. | | Deposit date: | 2006-10-24 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization.
Embo J., 27, 2008
|
|
2QEY
 
 | | Rat cytosolic PEPCK in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Sullivan, S.M, Holyoak, T. | | Deposit date: | 2007-06-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of rat cytosolic PEPCK: insight into the mechanism of phosphorylation and decarboxylation of oxaloacetic acid.
Biochemistry, 46, 2007
|
|
2LE0
 
 | | PARP BRCT Domain | | Descriptor: | Poly [ADP-ribose] polymerase 1 | | Authors: | Mueller, G, Loeffler, P, Cuneo, M, Derose, E, London, R. | | Deposit date: | 2011-06-03 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the PARP-1 BRCT domain.
Bmc Struct.Biol., 11, 2011
|
|
4BZ8
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1038 | | Descriptor: | (2R)-2-methyl-3-oxo-4H-1,4-benzothiazine-6-carbohydroxamic acid, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
1WCC
 
 | | Screening for fragment binding by X-ray crystallography | | Descriptor: | 2-AMINO-6-CHLOROPYRAZINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Cleasby, A, O'Reilly, M, Hartshorn, M.J, Murray, C.W, Tickle, I.J, Jhoti, H, Frederickson, M. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
4BZ7
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ9
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1075 | | Descriptor: | 3-chlorobenzothiophene-2-carbohydroxamic acid, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ5
 
 | | Crystal structure of Schistosoma mansoni HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
7E34
 
 | | Crystal structure of SUN1-Speedy A-CDK2 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, SUN domain-containing protein 1, ... | | Authors: | Chen, Y, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Nat Commun, 12, 2021
|
|
2NVU
 
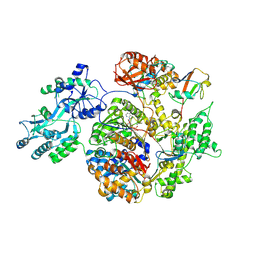 | | Structure of APPBP1-UBA3~NEDD8-NEDD8-MgATP-Ubc12(C111A), a trapped ubiquitin-like protein activation complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera, ... | | Authors: | Huang, D.T, Hunt, H.W, Zhuang, M, Ohi, M.D, Holton, J.M, Schulman, B.A. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Nature, 445, 2007
|
|
7C9P
 
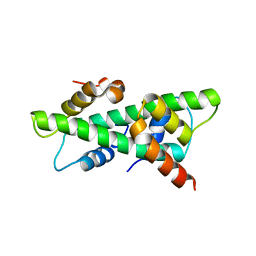 | | Crystal structure of rice histone-fold dimer GHD8/OsNF-YC2 | | Descriptor: | Nuclear transcription factor Y subunit B-11, Nuclear transcription factor Y subunit C-2 | | Authors: | Shen, C, Liu, H, Guan, Z, Xing, Y, Yin, P. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering.
Plant Cell, 32, 2020
|
|
4BZ6
 
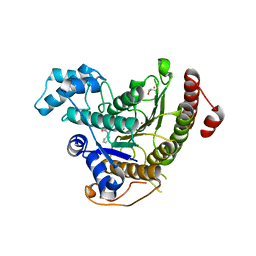 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with SAHA | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
1YWT
 
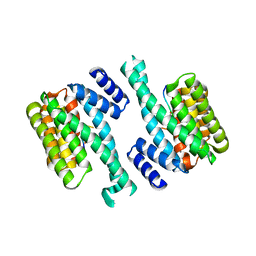 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
