5MST
 
 | |
3AG9
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2OUU
 
 | | crystal structure of PDE10A2 mutant D674A in complex with cGMP | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7SX5
 
 | | Crystal structure of ligase I with nick duplexes containing mismatch A:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
2OUS
 
 | | crystal structure of PDE10A2 mutant D674A | | Descriptor: | MAGNESIUM ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7SXE
 
 | | Crystal structure of ligase I with nick duplexes containing cognate G:T | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA chain 1, DNA chain 2, ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7SUM
 
 | | Crystal structure of human ligase I with nick duplexes containing cognate A:T | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, DNA(5'-*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*A-3'), ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
3AGM
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-670 | | Descriptor: | N~2~-{8-OXO-8-[4-(9H-PURIN-6-YL)PIPERAZIN-1-YL]OCTANOYL}-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGINYL-D-ARGININAMIDE, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2OUQ
 
 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OUV
 
 | | crystal structure of pde10a2 mutant of D564N | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3AGL
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1039 | | Descriptor: | (10R,20R,23R)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamido propyl)-10-methyl-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2OUP
 
 | | crystal structure of PDE10A | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8HLK
 
 | | Structure of McyB-C1A1 complexed with L-Leu and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, LEUCINE, Microcystin synthetase B (Fragment) | | Authors: | Peng, Y.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.
Structure, 32, 2024
|
|
3EHA
 
 | |
6YE1
 
 | | Human Ecto-5'-nucleotidase (CD73) in complex with the AMPCP derivative A894 (compound 2n in publication) in the closed form (crystal form IV) | | Descriptor: | 5'-nucleotidase, ZINC ION, [(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-(cyclopentylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxymethylphosphonic acid | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of Potent and Selective Methylenephosphonic Acid CD73 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6YE2
 
 | |
8CP4
 
 | | [4Fe-4S] cluster containing LarE in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
5N82
 
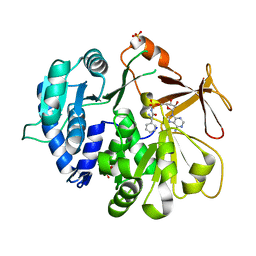 | | Crystal structure of an engineered TycA variant in complex with an beta-Phe-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.L, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
3SE5
 
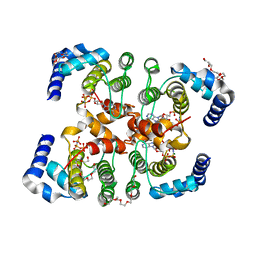 | | Fic protein from NEISSERIA MENINGITIDIS mutant delta8 in complex with AMPPNP | | Descriptor: | Cell filamentation protein Fic-related protein, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Goepfert, A, Stanger, F, Schirmer, T. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins.
Nature, 482, 2012
|
|
5N81
 
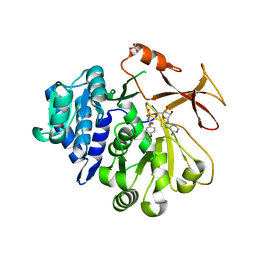 | | Crystal structure of an engineered TycA variant in complex with an O-propargyl-beta-Tyr-AMP analog | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Tyrocidine synthase 1, ... | | Authors: | Niquille, D.L, Hansen, D.A, Mori, T, Fercher, D, Kries, H, Hilvert, D. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonribosomal biosynthesis of backbone-modified peptides.
Nat Chem, 10, 2018
|
|
4DIN
 
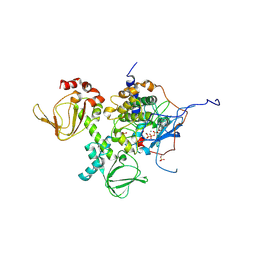 | | Novel Localization and Quaternary Structure of the PKA RI beta Holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Ilouz, R, Bubis, J, Wu, J, Yim, Y.Y, Deal, M.S, Kornev, A.P, Ma, Y, Blumenthal, D.K, Taylor, S.S. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Localization and quaternary structure of the PKA RI Beta holoenzyme
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UDT
 
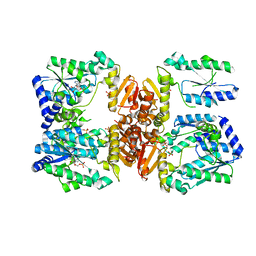 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lactate racemization operon protein LarE, PHOSPHATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1UXV
 
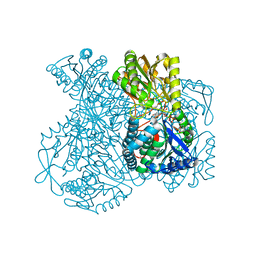 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXU
 
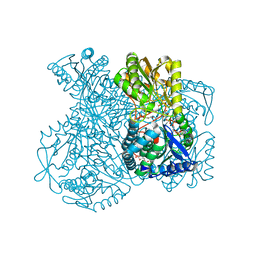 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1UXN
 
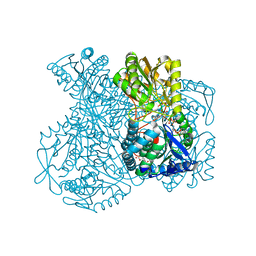 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
