2QFY
 
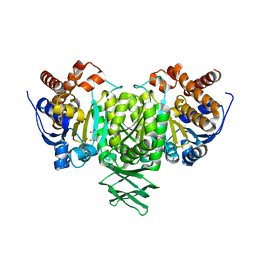 | |
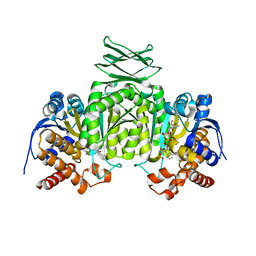
2QFV
 
 | |
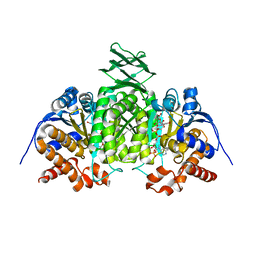
2QFX
 
 | |
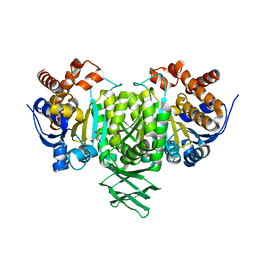
2QFW
 
 | |
8HLK
 
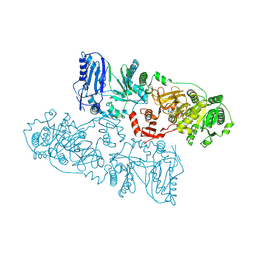 | |
8JBR
 
 | | Structure of McyA2-CAPCP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, McyA protein | | Authors: | Peng, Y.J. | | Deposit date: | 2023-05-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of McyA2-CAPCP
To Be Published
|
|
8GUF
 
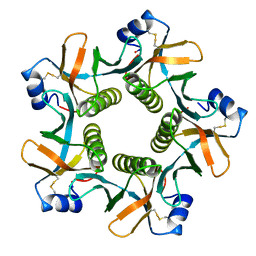 | |
8HJA
 
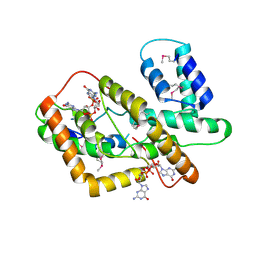 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | Authors: | Zeng, X, Peng, Y.J. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
