6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
1T03
 
 | | HIV-1 reverse transcriptase crosslinked to tenofovir terminated template-primer (complex P) | | Descriptor: | MAGNESIUM ION, POL polyprotein, Synthetic oligonucleotide primer, ... | | Authors: | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | Deposit date: | 2004-04-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
3BT2
 
 | | Structure of urokinase receptor, urokinase and vitronectin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | Authors: | Huang, M. | | Deposit date: | 2007-12-27 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
6H2Y
 
 | | human Fab 1E6 bound to fHbp variant 3 from Neisseria meningitidis serogroup B | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Veggi, D, Bianchi, F, Cozzi, R, Malito, E, Bottomley, M.J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cocrystal structure of meningococcal factor H binding protein variant 3 reveals a new crossprotective epitope recognized by human mAb 1E6.
Faseb J., 33, 2019
|
|
8HEG
 
 | | Pentamer of FMDV (A/TUR/14/98) in complex with M3F | | Descriptor: | M3F, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HBJ
 
 | | cocktail of FMDV (A/TUR/14/98) in complex with M678F and M688F | | Descriptor: | M678F nab, M688F nab, VP1 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HBG
 
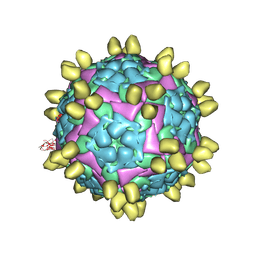 | | FMDV (A/TUR/14/98) in complex with M678F | | Descriptor: | M678F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
8HBI
 
 | | FMDV (A/TUR/14/98) in complex with M688F | | Descriptor: | M688F nab, VP1 of capsid protein, VP2 of capsid protein, ... | | Authors: | Li, H.Z, Dong, H, Liu, P. | | Deposit date: | 2022-10-28 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Foot-and-mouth disease virus antigenic landscape and reduced immunogenicity elucidated in atomic detail.
Nat Commun, 15, 2024
|
|
4M1D
 
 | |
1ACY
 
 | |
6Z6V
 
 | | Globular head of C1q in complex with the nanobody C1qNb75 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, ... | | Authors: | Laursen, N.S, Andersen, G.R. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Functional and Structural Characterization of a Potent C1q Inhibitor Targeting the Classical Pathway of the Complement System.
Front Immunol, 11, 2020
|
|
8CIF
 
 | | Bovine naive ultralong antibody AbD08 collected at 293K | | Descriptor: | Heavy chain, Light chain | | Authors: | Clarke, J.D, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
8DT8
 
 | | LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LM18 nanobody, Nb136 nanobody, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1XGY
 
 | | Crystal Structure of Anti-Meta I Rhodopsin Fab Fragment K42-41L | | Descriptor: | K42-41L Fab Heavy Chain, K42-41L Fab Light Chain, Rhodopsin Epitope Mimetic Peptide | | Authors: | Piscitelli, C.L, Angel, T.E, Bailey, B.W, Lawerence, C.M. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Equilibrium between metarhodopsin-I and metarhodopsin-II is dependent on the conformation of the third cytoplasmic loop.
J.Biol.Chem., 281, 2006
|
|
3BGF
 
 | |
6FRJ
 
 | | Crystal structure of scFv-SM3 in complex with APD-SeThrGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, APD-SeThr-RP, ... | | Authors: | Companon, I, Castro-Lopez, J, Escudero-Casao, M, Avenoza, A, Busto, J.H, Castillon, S, Jimenez-Barbero, J, Bernardes, G.J, Boutureira, O, Jimenez-Oses, G, Asensio, J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2018-02-16 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent Tumor-Associated Antigens: Modulation of Peptide Presentation by Single-Atom O/S or O/Se Substitutions at the Glycosidic Linkage.
J. Am. Chem. Soc., 141, 2019
|
|
6Y9B
 
 | |
3DNO
 
 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DNN
 
 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
8URF
 
 | |
2JB5
 
 | | Fab fragment in complex with small molecule hapten, crystal form-1 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
2JB6
 
 | | Fab fragment in complex with small molecule hapten, crystal form-2 | | Descriptor: | 2-{(1E,3Z,5E,7E)-7-[3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-1,3-DIHYDRO-2H-INDOL-2-YLIDENE]-4-METHYLHEPTA-1,3,5-TRIEN-1-YL}-3,3-DIMETHYL-5-SULFO-1-(2-SULFOETHYL)-3H-INDOLIUM, FAB FRAGMENT MOR03268 HEAVY CHAIN, FAB FRAGMENT MOR03268 LIGHT CHAIN | | Authors: | Hillig, R.C, Baesler, S, Malawski, G, Badock, V, Bahr, I, Schirner, M, Licha, K. | | Deposit date: | 2006-12-03 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fab Mor03268 Triggers Absorption Shift of a Diagnostic Dye Via Packaging in a Solvent-Shielded Fab Dimer Interface
J.Mol.Biol., 377, 2008
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
