6UMX
 
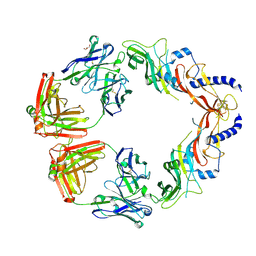 | | Structural basis for specific inhibition of extracellular activation of pro/latent myostatin by SRK-015 | | Descriptor: | GL29H4-16 Fab Heavy Chain,GL29H4-16 Fab Heavy Chain, GL29H4-16 Fab Light Chain,GL29H4-16 Fab Light Chain, GLYCEROL, ... | | Authors: | Dagbay, K.B, Treece, E, Streich Jr, F.C, Jackson, J.W, Faucette, R.R, Nikiforov, A, Lin, S.C, Bostion, C.J, Nicholls, S.B, Capili, A.D, Carven, G.J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of specific inhibition of extracellular activation of pro- or latent myostatin by the monoclonal antibody SRK-015.
J.Biol.Chem., 295, 2020
|
|
7KBB
 
 | |
7KBA
 
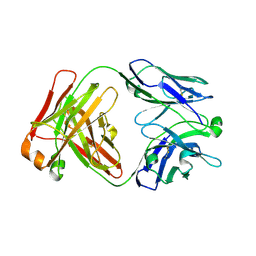 | |
6LCH
 
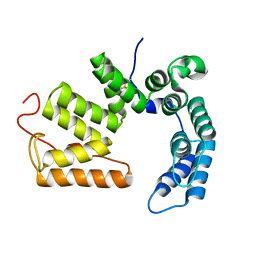 | | Pseudomonas aeruginosa 5086 (PA5086) | | Descriptor: | Sel1 repeat family protein | | Authors: | She, Z, Geng, Z. | | Deposit date: | 2019-11-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa T6SS PldB immunity proteins PA5086, PA5087 and PA5088 explains a novel stockpiling mechanism.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7M30
 
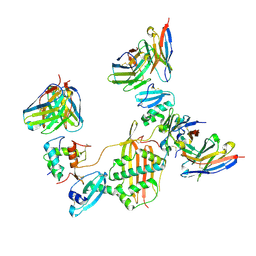 | |
7M1C
 
 | |
7LYV
 
 | |
7LYW
 
 | |
8VFK
 
 | | Crystal Structure of Delta 109-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRATE ANION, D-dopachrome decarboxylase, SODIUM ION | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFN
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 310K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG5
 
 | | Crystal Structure of V113N Variant of D-Dopachrome Tautomerase (D-DT) Bound with 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VDY
 
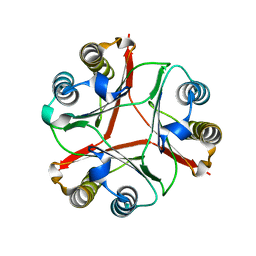 | | Crystal Structure of Delta 114-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VFL
 
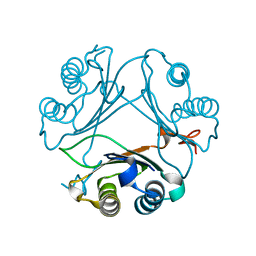 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 290K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
2NAB
 
 | | Nizp1-C2HR zinc finger structure | | Descriptor: | ZINC ION, Zinc finger protein 496 | | Authors: | Berardi, A, Quilici, G, Spiliotopoulos, D, Corral-Rodriguez, M, Martin, F, Degano, M, Tonon, G, Musco, G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: implications for Sotos syndrome.
Nucleic Acids Res., 44, 2016
|
|
2NAA
 
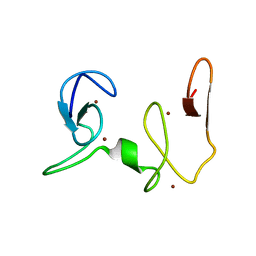 | | NSD1-PHD_5-C5HCH tandem domain structure | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, ZINC ION | | Authors: | Berardi, A, Quilici, G, Spiliotopoulos, D, Musco, G. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for PHDVC5HCHNSD1-C2HRNizp1 interaction: implications for Sotos syndrome.
Nucleic Acids Res., 44, 2016
|
|
7QAC
 
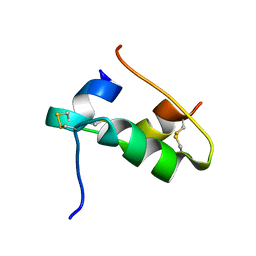 | | The T2 structure of polycrystalline cubic human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Karavassili, F, Triandafillidis, D.P, Valmas, A, Spiliopoulou, M, Fili, S, Kontou, P, Bowler, M.W, Von Dreele, R.B, Fitch, A, Margiolaki, I. | | Deposit date: | 2021-11-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | POWDER DIFFRACTION (2.29 Å) | | Cite: | The T 2 structure of polycrystalline cubic human insulin.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6FGV
 
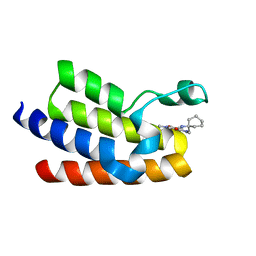 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGT
 
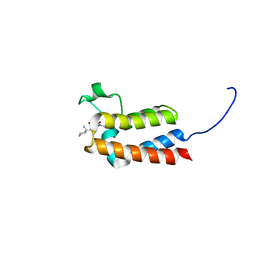 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGG
 
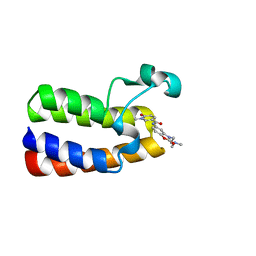 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 5 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[3-[2-(dimethylamino)ethyl]-2-oxidanylidene-1,3-benzoxazol-5-yl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGW
 
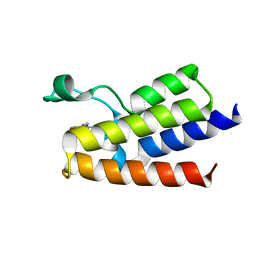 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 4 | | Descriptor: | 1-methyl-6-oxidanylidene-~{N}-(2-pyrrolidin-1-ylethyl)pyridine-3-carboxamide, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FH6
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-(2-azanylethyl)-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FG6
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-(2-azanylethyl)-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
