3V8I
 
 | |
1KBL
 
 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | AMMONIUM ION, PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C, Liu, S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1F4B
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F51
 
 | | A TRANSIENT INTERACTION BETWEEN TWO PHOSPHORELAY PROTEINS TRAPPED IN A CRYSTAL LATTICE REVEALS THE MECHANISM OF MOLECULAR RECOGNITION AND PHOSPHOTRANSFER IN SINGAL TRANSDUCTION | | Descriptor: | MAGNESIUM ION, SPORULATION INITIATION PHOSPHOTRANSFERASE B, SPORULATION INITIATION PHOSPHOTRANSFERASE F | | Authors: | Zapf, J, Sen, U, Madhusudan, M, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2000-06-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction.
Structure Fold.Des., 8, 2000
|
|
1U7N
 
 | | Crystal Structure of the fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Fatty acid/phospholipid synthesis protein plsX | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
1PSS
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
2ZW4
 
 | |
1PCR
 
 | | STRUCTURE OF THE PHOTOSYNTHETIC REACTION CENTRE FROM RHODOBACTER SPHAEROIDES AT 2.65 ANGSTROMS RESOLUTION: COFACTORS AND PROTEIN-COFACTOR INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Ermler, U, Fritzsch, G, Michel, H. | | Deposit date: | 1994-11-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions.
Structure, 2, 1994
|
|
2ZXV
 
 | | Crystal structure of putative acetyltransferase from T. thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHA1799 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
1F28
 
 | | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII BOUND TO DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Anderson, A.C, O'Neil, R.H, Surti, T.S, Stroud, R.M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking.
Chem.Biol., 8, 2001
|
|
4PVX
 
 | | Crystal structure of human FPPS in complex with [({4-[4-(cyclopropyloxy)phenyl]pyridin-2-yl}amino)methanediyl]bis(phosphonic acid) | | Descriptor: | Farnesyl pyrophosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rodionov, D, Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-03-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
3VJD
 
 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus | | Descriptor: | Dehydrosqualene synthase, L(+)-TARTARIC ACID | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
3VK5
 
 | | Crystal structure of MoeO5 in complex with its product FPG | | Descriptor: | (2R)-3-(phosphonooxy)-2-{[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}propanoic acid, MAGNESIUM ION, MoeO5 | | Authors: | Ren, F, Ko, T.-P, Huang, C.-H, Guo, R.-T. | | Deposit date: | 2011-11-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Insights into the mechanism of the antibiotic-synthesizing enzyme MoeO5 from crystal structures of different complexes
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1F4F
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH SP-722 | | Descriptor: | 4-[[GLUTAMIC ACID]-CARBONYL]-BENZENE-SULFONYL-D-PROLINE, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ZY1
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-830 | | Descriptor: | Dehydrosqualene synthase, dipotassium (2-oxo-2-{[3-(3-phenoxyphenyl)propyl]amino}ethyl)phosphonate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2009-01-10 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic results.
J.Med.Chem., 52, 2009
|
|
3VJC
 
 | | Crystal structure of the human squalene synthase in complex with zaragozic acid A | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Squalene synthase, ... | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
1TSX
 
 | | THYMIDYLATE SYNTHASE R179E MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1F4G
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH SP-876 | | Descriptor: | GLYCEROL, N-[4-[[GLUTAMIC ACID]-CARBONYL]-BENZENE-SULFONYL-D-PROLINYL]-3-AMINO-PROPANOIC ACID, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KUV
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL 5-BROMOTRYPTAMINE, MAGNESIUM ION, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
3VZZ
 
 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
1F75
 
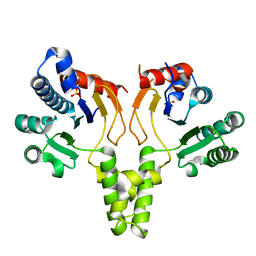 | | CRYSTAL STRUCTURE OF UNDECAPRENYL DIPHOSPHATE SYNTHASE FROM MICROCOCCUS LUTEUS B-P 26 | | Descriptor: | SULFATE ION, UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Fujihashi, M, Zhang, Y.-W, Higuchi, Y, Li, X.-Y, Koyama, T, Miki, K. | | Deposit date: | 2000-06-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3A27
 
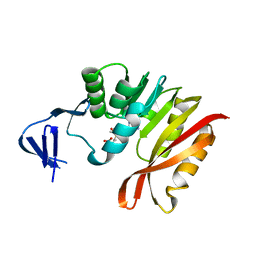 | | Crystal structure of M. jannaschii TYW2 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein MJ1557 | | Authors: | Umitsu, M, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4H5D
 
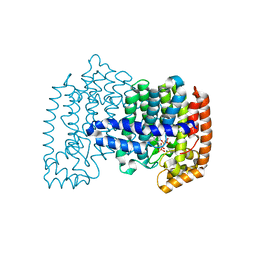 | | Crystal structure of human FPPS in ternary complex with YS0470 and inorganic pyrophosphate | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-09-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Ternary complex structures of human farnesyl pyrophosphate synthase bound with a novel inhibitor and secondary ligands provide insights into the molecular details of the enzyme's active site closure.
Bmc Struct.Biol., 12, 2012
|
|
2ZXU
 
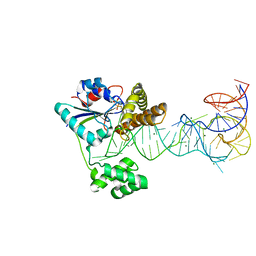 | | Crystal structure of tRNA modification enzyme MiaA in the complex with tRNA(Phe) and DMASPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, tRNA delta(2)-isopentenylpyrophosphate transferase, ... | | Authors: | Sakai, J, Yao, M, Chimnaronk, S, Tanaka, I. | | Deposit date: | 2009-01-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon
Biochemistry, 48, 2009
|
|
1F4C
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE COVALENTLY MODIFIED AT C146 WITH N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL | | Descriptor: | GLYCEROL, N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
