1T05
 
 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | Descriptor: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | Authors: | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | Deposit date: | 2004-04-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
6J1Z
 
 | | WWP2 semi-open conformation | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Liu, Z.H. | | Deposit date: | 2018-12-30 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.
Nat Commun, 10, 2019
|
|
1T52
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T51
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
6IZ6
 
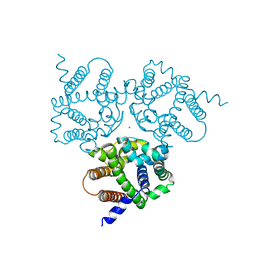 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | CALCIUM ION, Trimeric intracellular cation channel type B-B | | Authors: | Liu, X.L, Wang, X.H, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZF
 
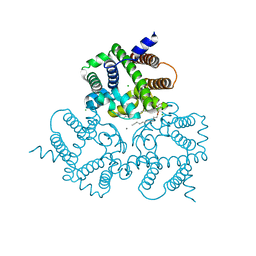 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1TAT
 
 | |
6J5I
 
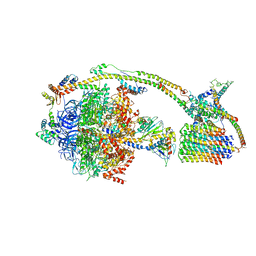 | | Cryo-EM structure of the mammalian DP-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
1TBY
 
 | |
1T8I
 
 | | Human DNA Topoisomerase I (70 Kda) In Complex With The Poison Camptothecin and Covalent Complex With A 22 Base Pair DNA Duplex | | Descriptor: | 4-ETHYL-4-HYDROXY-1,12-DIHYDRO-4H-2-OXA-6,12A-DIAZA-DIBENZO[B,H]FLUORENE-3,13-DIONE, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', ... | | Authors: | Staker, B.L, Feese, M.D, Cushman, M, Pommier, Y, Zembower, D, Stewart, L, Burgin, A.B. | | Deposit date: | 2004-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex
J.Med.Chem., 48, 2005
|
|
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
6J8M
 
 | | Low-dose structure of bovine heart cytochrome c oxidase in the fully oxidized state determined using 30 keV X-ray | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ueno, G, Shimada, A, Yamashita, E, Hasegawa, K, Kumasaka, T, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T, Yamamoto, M. | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays.
J.Synchrotron Radiat., 26, 2019
|
|
1TLO
 
 | | High resolution crystal structure of calpain I protease core in complex with E64 | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
8GUS
 
 | | Cryo-EM structure of HU-CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
1SU5
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
6J20
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
6JB1
 
 | | Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ATP-binding cassette sub-family C member 8 isoform X2, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
1SW7
 
 | | Triosephosphate isomerase from Gallus gallus, loop 6 mutant K174N, T175S, A176S | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
6J31
 
 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
8GLV
 
 | |
8GHV
 
 | | Cannabinoid Receptor 1-G Protein Complex | | Descriptor: | (5Z,8Z,11Z,13S,14Z)-N-[(2R)-1-hydroxypropan-2-yl]-13-methylicosa-5,8,11,14-tetraenamide, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, Robertson, M.J, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2023-03-12 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for activation of CB1 by an endocannabinoid analog.
Nat Commun, 14, 2023
|
|
8GUQ
 
 | | Cryo-EM structure of CB2-G protein complex | | Descriptor: | Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wu, L.J, Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-05-10 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of selective cannabinoid CB 2 receptor activation.
Nat Commun, 14, 2023
|
|
6J54
 
 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
1SZM
 
 | | DUAL BINDING MODE OF BISINDOLYLMALEIMIDE 2 TO PROTEIN KINASE A (PKA) | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Gassel, M, Breitenlechner, C.B, Koenig, N, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The protein kinase C inhibitor bisindolyl maleimide 2 binds with reversed orientations to different conformations of protein kinase a.
J.Biol.Chem., 279, 2004
|
|
