8IKL
 
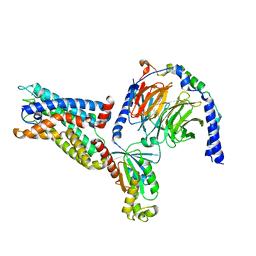 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8IXZ
 
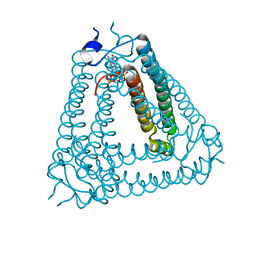 | |
8IY1
 
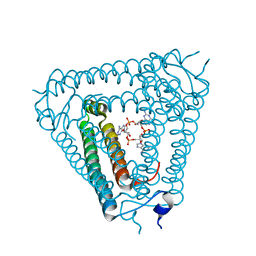 | | Structure of Acb2 complexed with cAAG | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8IY0
 
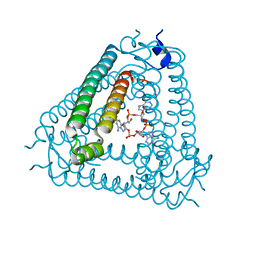 | | Structure of Acb2 complexed with cAAA | | Descriptor: | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, p26 | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IY2
 
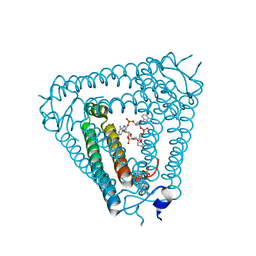 | | Structure of Acb2 complexed with 3',3'-cGAMP and cAAA | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-3'-MONOPHOSPHATE, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate, ... | | Authors: | Cao, X.L, Xiao, Y, Feng, Y. | | Deposit date: | 2023-04-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Phage anti-CBASS protein simultaneously sequesters cyclic trinucleotides and dinucleotides.
Mol.Cell, 84, 2024
|
|
8IUF
 
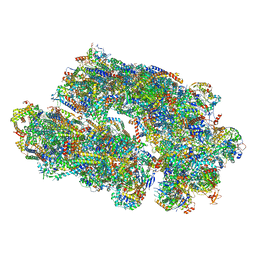 | | Cryo-EM structure of Euglena gracilis super-complex I+III2+IV, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
1GVH
 
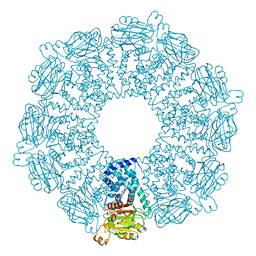 | | The X-ray structure of ferric Escherichia coli flavohemoglobin reveals an unespected geometry of the distal heme pocket | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ilari, A, Johnson, K.A, Bonamore, A, Farina, A, Boffi, A. | | Deposit date: | 2002-02-13 | | Release date: | 2002-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The X-Ray Structure of Ferric Escherichia Coli Flavohemoglobin Reveals an Unexpected Geometry of the Distal Heme Pocket
J.Biol.Chem., 277, 2002
|
|
1HBG
 
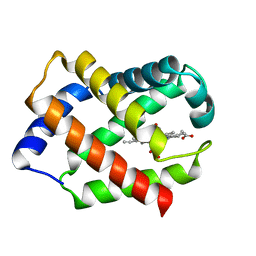 | | GLYCERA DIBRANCHIATA HEMOGLOBIN. STRUCTURE AND REFINEMENT AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (CARBONMONOXY), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arents, G.A, Braden, B.C, Padlan, E.A, Love, W.E. | | Deposit date: | 1991-02-11 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Glycera dibranchiata hemoglobin. Structure and refinement at 1.5 A resolution.
J.Mol.Biol., 210, 1989
|
|
1H25
 
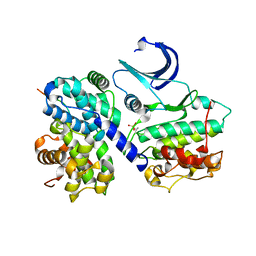 | | CDK2/Cyclin A in complex with an 11-residue recruitment peptide from retinoblastoma-associated protein | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, RETINOBLASTOMA-ASSOCIATED PROTEIN | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
6QHK
 
 | |
8FY5
 
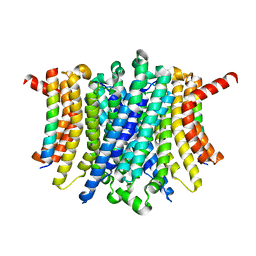 | | Human TMEM175-LAMP1 full-length complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
8FYF
 
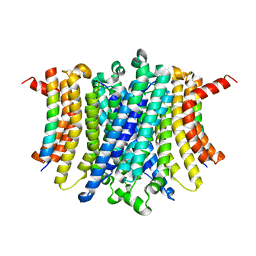 | | Human TMEM175-LAMP1 transmembrane domain only complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
8GAT
 
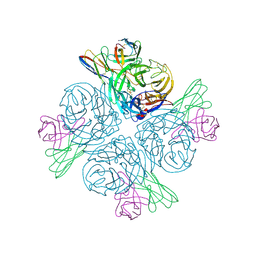 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAV
 
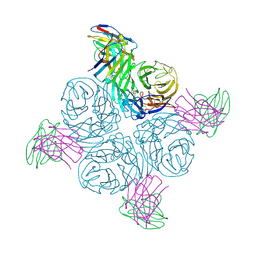 | | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab NDS.3, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAU
 
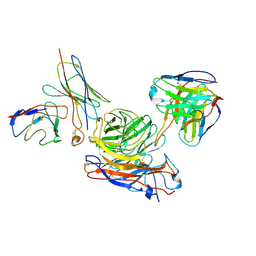 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
6H6N
 
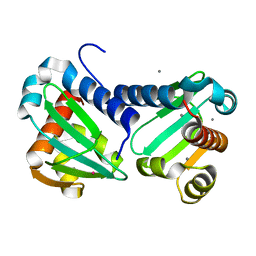 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | CALCIUM ION, TERBIUM(III) ION, Ubiquinone biosynthesis protein UbiJ | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6GZ4
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-2 (TI-POST-2) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6H6O
 
 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6RXU
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6GZ3
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-1 (TI-POST-1) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8GOC
 
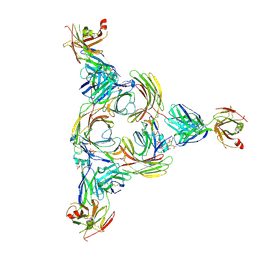 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Vasopressin V2 receptor, V2R | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GO8
 
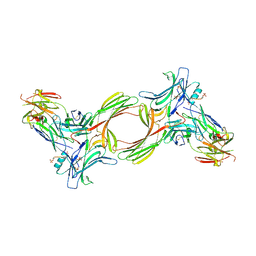 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
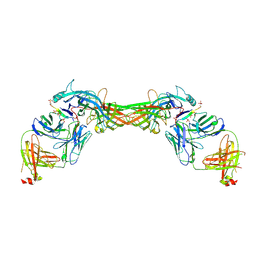 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
