6I0K
 
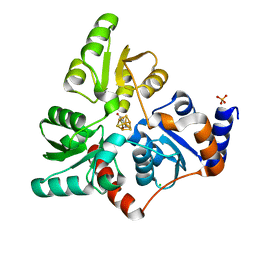 | | Structure of quinolinate synthase in complex with 4-mercaptophthalic acid | | Descriptor: | 4-mercaptoidenecyclohexa-2,5-diene-1,2-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
8Q2R
 
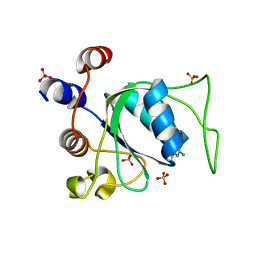 | | Crystal structure of YTHDC1 in complex with Compound 3 (ZA_431) | | Descriptor: | 5-chloranyl-~{N},3-dimethyl-1~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Zalesak, F, Li, Y, Caflisch, A. | | Deposit date: | 2023-08-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of a Potent and Selective YTHDC1 Ligand.
J.Med.Chem., 67, 2024
|
|
8D95
 
 | |
8T5M
 
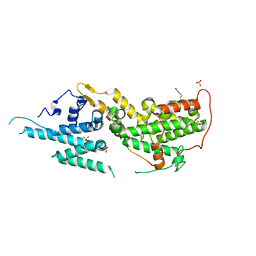 | | SOS2 crystal structure with fragment bound (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8ZFO
 
 | |
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
8Y7L
 
 | |
8ZFR
 
 | |
8C3X
 
 | |
8ZFP
 
 | |
5LO9
 
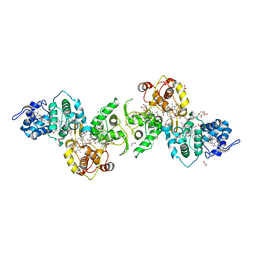 | | Thiosulfate dehydrogenase (TsdBA) from Marichromatium purpuratum - "as isolated" form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome C, ... | | Authors: | Brito, J.A, Kurth, J.M, Reuter, J, Flegler, A, Koch, T, Franke, T, Klein, E, Rowe, S, Butt, J.N, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase.
J.Biol.Chem., 291, 2016
|
|
5LPW
 
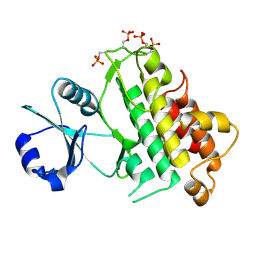 | |
6TS9
 
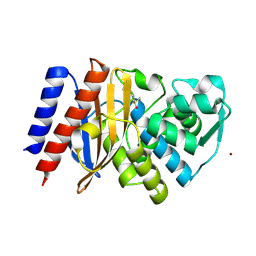 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
6R85
 
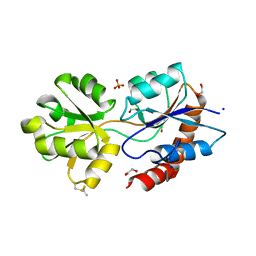 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6HYS
 
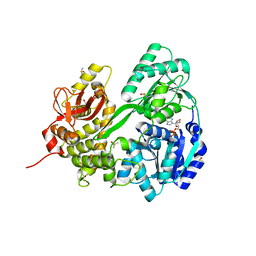 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.6 angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
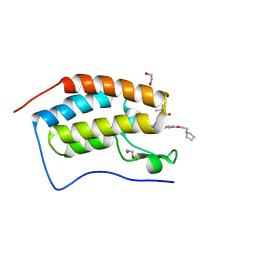
8EAD
 
 | |
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4RVN
 
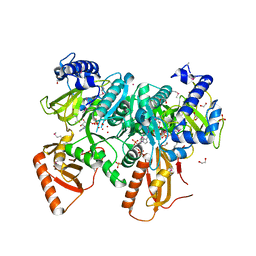 | |
7BYD
 
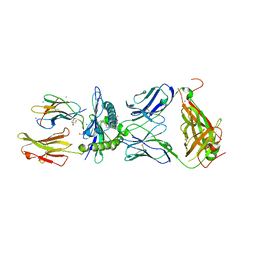 | | Crystal structure of SN45 TCR in complex with lipopeptide-bound Mamu-B*05104 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, B protein, ... | | Authors: | Morita, D, Sugita, M, Iwashita, C. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.80003262 Å) | | Cite: | Crystal structure of the ternary complex of TCR, MHC class I and lipopeptides.
Int.Immunol., 32, 2020
|
|
8C1A
 
 | |
8CK0
 
 | |
6RDJ
 
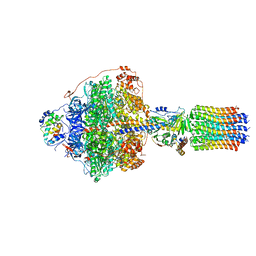 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5NQ1
 
 | | Porcine (Sus scrofa) Major Histocompatibility Complex, class I, with human beta2 micro globulin, presenting DFEREGYSL | | Descriptor: | 1,2-ETHANEDIOL, ASP-PHE-GLU-ARG-GLU-GLY-TYR-SER-LEU, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Tungatt, K, Sewell, A.K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Induction of influenza-specific local CD8 T-cells in the respiratory tract after aerosol delivery of vaccine antigen or virus in the Babraham inbred pig.
Plos Pathog., 14, 2018
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
6XIX
 
 | |
