6A53
 
 | | Crystal structure of DddK | | Descriptor: | MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6LHP
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
5U68
 
 | |
6LHO
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
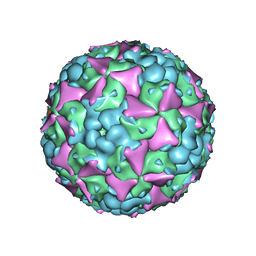 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
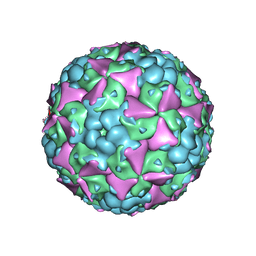 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
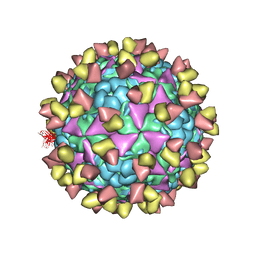 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHT
 
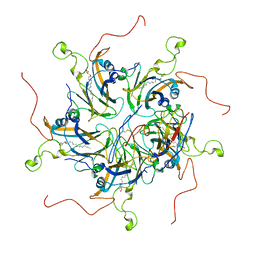 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6MEL
 
 | | Succinyl-CoA synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
6LU5
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-25 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86527729 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-26 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.970063 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7K6R
 
 | |
7K6S
 
 | | Crystal structure of the bromodomain (BD) of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to Compound 4 (SKT1174) | | Descriptor: | (7-amino-3,4-dihydroquinolin-1(2H)-yl)(cyclopropyl)methanone, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay development.
Org.Biomol.Chem., 18, 2020
|
|
7KDW
 
 | |
6AZE
 
 | |
5UZ9
 
 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LP0
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRO
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5VP5
 
 | |
5W49
 
 | | The crystal structure of human S-adenosylhomocysteine hydrolase (AHCY) bound to oxadiazole inhibitor | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[(3R)-3-{4-[(3-methoxyphenyl)amino]-6-methylpyridin-2-yl}pyrrolidin-1-yl]methanone, 1,2-ETHANEDIOL, Adenosylhomocysteinase, ... | | Authors: | Dougan, D.R, Lawson, J.D, Lane, W. | | Deposit date: | 2017-06-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of AHCY inhibitors using novel high-throughput mass spectrometry.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
7KDZ
 
 | |
